Structural characterization of GASDALIE Fc bound to the activating Fc receptor FcγRIIIa
- PMID: 26850169
- PMCID: PMC4769027
- DOI: 10.1016/j.jsb.2016.02.001
Structural characterization of GASDALIE Fc bound to the activating Fc receptor FcγRIIIa
Abstract
The Fc region of Immunoglobulin G (IgG) initiates inflammatory responses such as antibody-dependent cell-mediated cytotoxicity (ADCC) through binding to activating Fc receptors (FcγRI, FcγRIIa, FcγRIIIa). These receptors are expressed on the surface of immune cells including macrophages, dendritic cells, and natural killer cells. An inhibitory receptor, FcγRIIb, is expressed on macrophages and other myeloid leukocytes simultaneously with the activating receptor FcγRIIa, thereby setting a threshold for cell activation. The affinity of IgG Fc for binding activating Fc receptors depends on IgG subclass and the composition of N-linked glycans attached to a conserved asparagine in the Fc CH2 domain. For example, Fc regions with afucosylated glycans bind more tightly to FcγRIIIa than fucosylated Fc, and afucosylated Fcs exhibit enhanced ADCC activity in vivo and in vitro. Enhanced pro-inflammatory responses have also been seen for Fc regions with amino acid substitutions. GASDALIE Fc is an Fc mutant (G236A/S239D/A330L/I332E) that exhibits a higher affinity for FcγRIIIa and increased effector functions in vivo compared to wild-type Fc. To explore its altered functions, we compared the affinities of GASDALIE and wild-type Fc for activating and inhibitory FcγRs. We also determined the crystal structure of GASDALIE Fc alone and bound to FcγRIIIa. The overall structure of GASDALIE Fc alone was similar to wild-type Fc structures, however, increased electrostatic interactions in the GASDALIE Fc:FcγRIIIa interface compared with other Fc:FcγR structures suggest a mechanism for the increased affinity of GASDALIE Fc for FcγRIIIa.
Keywords: Affinity; Fc receptors; Fc–FcR structure; GASDALIE Fc; IgG Fc; SDALIE Fc.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



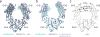
References
-
- Ackerman ME, Nimmerjahn F. Antibody Fc linking adaptive and innate immunity. Amsterdam; Burlington: Elsevier Science; 2014. p. 1. online resource (xii, 363 pages)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

