Auxin-Independent NAC Pathway Acts in Response to Explant-Specific Wounding and Promotes Root Tip Emergence during de Novo Root Organogenesis in Arabidopsis
- PMID: 26850273
- PMCID: PMC4825153
- DOI: 10.1104/pp.15.01733
Auxin-Independent NAC Pathway Acts in Response to Explant-Specific Wounding and Promotes Root Tip Emergence during de Novo Root Organogenesis in Arabidopsis
Abstract
Plants have powerful regenerative abilities that allow them to recover from damage and survive in nature. De novo organogenesis is one type of plant regeneration in which adventitious roots and shoots are produced from wounded and detached organs. By studying de novo root organogenesis using leaf explants of Arabidopsis (Arabidopsis thaliana), we previously suggested that wounding is the first event that provides signals to trigger the whole regenerative process. However, our knowledge of the role of wounding in regeneration remains limited. In this study, we show that wounding not only triggers the auxin-mediated fate transition of regeneration-competent cells, but also induces the NAC pathway for root tip emergence. The NAC1 transcription factor gene was specifically expressed in response to wounding in the leaf explant, but not in the wounded leaf residue of the source plant. Inhibition of the NAC1 pathway severely affected the emergence of adventitious root tips. However, the NAC1 pathway functioned independently of auxin-mediated cell fate transition and regulates expression of CEP genes, which encode proteins that might have a role in degradation of extensin proteins in the cell wall. Overall, our results suggest that wounding has multiple roles in de novo root organogenesis and that NAC1 acts as one downstream branch in regulating the cellular environment for organ emergence.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures
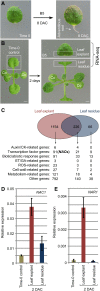

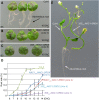
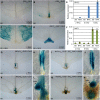
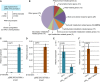


References
-
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 - PMC - PubMed
-
- Bostock RM. (1989) Perspectives on wound healing in resistance to pathogens. Annu Rev Phytopathol 27: 343–371
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

