High-Throughput Single-Cell Cultivation on Microfluidic Streak Plates
- PMID: 26850294
- PMCID: PMC4807504
- DOI: 10.1128/AEM.03588-15
High-Throughput Single-Cell Cultivation on Microfluidic Streak Plates
Abstract
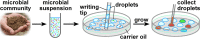
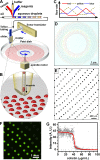
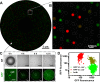
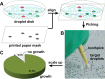
This paper describes the microfluidic streak plate (MSP), a facile method for high-throughput microbial cell separation and cultivation in nanoliter sessile droplets. The MSP method builds upon the conventional streak plate technique by using microfluidic devices to generate nanoliter droplets that can be streaked manually or robotically onto petri dishes prefilled with carrier oil for cultivation of single cells. In addition, chemical gradients could be encoded in the droplet array for comprehensive dose-response analysis. The MSP method was validated by using single-cell isolation of Escherichia coli and antimicrobial susceptibility testing of Pseudomonas aeruginosa PAO1. The robustness of the MSP work flow was demonstrated by cultivating a soil community that degrades polycyclic aromatic hydrocarbons. Cultivation in droplets enabled detection of the richest species diversity with better coverage of rare species. Moreover, isolation and cultivation of bacterial strains by MSP led to the discovery of several species with high degradation efficiency, including four Mycobacterium isolates and a previously unknown fluoranthene-degrading Blastococcus species.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Dore J, Ehrlich SD, Bork P, Wang J. 2014. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841. doi: 10.1038/nbt.2942. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

