Simultaneous Rapid Detection and Serotyping of Cronobacter sakazakii Serotypes O1, O2, and O3 by Using Specific Monoclonal Antibodies
- PMID: 26850303
- PMCID: PMC4959477
- DOI: 10.1128/AEM.04016-15
Simultaneous Rapid Detection and Serotyping of Cronobacter sakazakii Serotypes O1, O2, and O3 by Using Specific Monoclonal Antibodies
Abstract
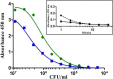
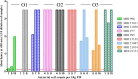
Cronobacter sakazakii is a foodborne pathogen associated with rare but often lethal infections in neonates. Powdered infant formula (PIF) represents the most frequent source of infection. Out of the identified serotypes (O1 to O7), O1, O2, and O3 are often isolated from clinical and PIF samples. Serotype-specific monoclonal antibodies (MAbs) suitable for application in enzyme immunoassays (EIAs) for the rapid detection of C. sakazakii have not yet been developed. In this study, we created specific MAbs with the ability to bind toC. sakazakii of serotypes O1, O2, and O3. Characterization by indirect EIAs, immunofluorescence, motility assays, and immunoblotting identified lipopolysaccharide (LPS) and exopolysaccharide (EPS) as the antigenic determinants of the MAbs. The established sandwich EIAs were highly sensitive and were able to detect between 2 × 10(3)and 9 × 10(6)CFU/ml. Inclusivity tests confirmed that 93% of serotype O1 strains, 100% of O2 strains, and 87% of O3 strains were detected at low cell counts. No cross-reactivity with >100 strains of Cronobacter spp. and other Enterobacter iaceae was observed, except for that with C. sakazakii serotype O3 and Cronobacter muytjensii serotype O1. Moreover, the sandwich EIAs detected C. sakazakii in PIF samples artificially contaminated with 1 to 10 bacterial cells per 10 g of sample after 15 h of preenrichment. The use of these serotype-specific MAbs not only allows the reliable detection of C. sakazakii strains but also enables simultaneous serotyping in a simple sandwich EIA method.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Multiplexed Lateral Flow Test for Detection and Differentiation of Cronobacter sakazakii Serotypes O1 and O2.Front Microbiol. 2017 Sep 20;8:1826. doi: 10.3389/fmicb.2017.01826. eCollection 2017. Front Microbiol. 2017. PMID: 28979257 Free PMC article.
-
Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes.Appl Environ Microbiol. 2012 Jun;78(11):3966-74. doi: 10.1128/AEM.07825-11. Epub 2012 Mar 23. Appl Environ Microbiol. 2012. PMID: 22447597 Free PMC article.
-
Genotyping and Source Tracking of Cronobacter sakazakii and C. malonaticus Isolates from Powdered Infant Formula and an Infant Formula Production Factory in China.Appl Environ Microbiol. 2015 Aug 15;81(16):5430-9. doi: 10.1128/AEM.01390-15. Epub 2015 Jun 5. Appl Environ Microbiol. 2015. PMID: 26048942 Free PMC article.
-
Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium.J Appl Microbiol. 2012 Jul;113(1):1-15. doi: 10.1111/j.1365-2672.2012.05281.x. Epub 2012 Apr 11. J Appl Microbiol. 2012. PMID: 22420458 Review.
-
Trending biocontrol strategies against Cronobacter sakazakii: A recent updated review.Food Res Int. 2020 Nov;137:109385. doi: 10.1016/j.foodres.2020.109385. Epub 2020 Jun 5. Food Res Int. 2020. PMID: 33233087 Review.
Cited by
-
Multiplexed Lateral Flow Test for Detection and Differentiation of Cronobacter sakazakii Serotypes O1 and O2.Front Microbiol. 2017 Sep 20;8:1826. doi: 10.3389/fmicb.2017.01826. eCollection 2017. Front Microbiol. 2017. PMID: 28979257 Free PMC article.
-
Molecular and Biological Characteristics of the Cronobacter Genus Bacteria Isolated from the Dry Infant Food Products.Bull Exp Biol Med. 2025 May;179(1):50-53. doi: 10.1007/s10517-025-06440-9. Epub 2025 Jul 19. Bull Exp Biol Med. 2025. PMID: 40682651
-
Voltammetric sandwich immunoassay for Cronobacter sakazakii using a screen-printed carbon electrode modified with horseradish peroxidase, reduced graphene oxide, thionine and gold nanoparticles.Mikrochim Acta. 2017 Dec 9;185(1):45. doi: 10.1007/s00604-017-2572-x. Mikrochim Acta. 2017. PMID: 29594632
References
-
- Lai KK. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113–122. - PubMed
-
- Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, Fanning S, Stephan R, Joosten H. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov., and descriptions of Cronobacter sakazakii comb. nov., Cronobacter sakazakii subsp. sakazakii comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol 7:64. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

