Insights into Coupled Folding and Binding Mechanisms from Kinetic Studies
- PMID: 26851275
- PMCID: PMC4807256
- DOI: 10.1074/jbc.R115.692715
Insights into Coupled Folding and Binding Mechanisms from Kinetic Studies
Abstract
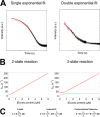
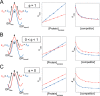
Intrinsically disordered proteins (IDPs) are characterized by a lack of persistent structure. Since their identification more than a decade ago, many questions regarding their functional relevance and interaction mechanisms remain unanswered. Although most experiments have taken equilibrium and structural perspectives, fewer studies have investigated the kinetics of their interactions. Here we review and highlight the type of information that can be gained from kinetic studies. In particular, we show how kinetic studies of coupled folding and binding reactions, an important class of signaling event, are needed to determine mechanisms.
Keywords: IDP; biophysics; coupled folding and binding; electrostatics; kinetics; phi-value; protein dynamic; protein electrostatics; protein folding; protein-protein interactions; residual structure; signaling.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

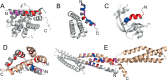
Similar articles
-
Folding and binding pathways of BH3-only proteins are encoded within their intrinsically disordered sequence, not templated by partner proteins.J Biol Chem. 2018 Jun 22;293(25):9718-9723. doi: 10.1074/jbc.RA118.002791. Epub 2018 May 1. J Biol Chem. 2018. PMID: 29716994 Free PMC article.
-
Mapping the transition state for a binding reaction between ancient intrinsically disordered proteins.J Biol Chem. 2020 Dec 18;295(51):17698-17712. doi: 10.1074/jbc.RA120.015645. J Biol Chem. 2020. PMID: 33454008 Free PMC article.
-
A structurally heterogeneous transition state underlies coupled binding and folding of disordered proteins.J Biol Chem. 2019 Jan 25;294(4):1230-1239. doi: 10.1074/jbc.RA118.005854. Epub 2018 Dec 4. J Biol Chem. 2019. PMID: 30514761 Free PMC article.
-
Role of Intrinsic Protein Disorder in the Function and Interactions of the Transcriptional Coactivators CREB-binding Protein (CBP) and p300.J Biol Chem. 2016 Mar 25;291(13):6714-22. doi: 10.1074/jbc.R115.692020. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851278 Free PMC article. Review.
-
Expanding the Range of Protein Function at the Far End of the Order-Structure Continuum.J Biol Chem. 2016 Mar 25;291(13):6706-13. doi: 10.1074/jbc.R115.692590. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851282 Free PMC article. Review.
Cited by
-
Site-Specific Polymer Attachment to HR2 Peptide Fusion Inhibitors against HIV-1 Decreases Binding Association Rates and Dissociation Rates Rather Than Binding Affinity.Bioconjug Chem. 2017 Mar 15;28(3):701-712. doi: 10.1021/acs.bioconjchem.6b00540. Epub 2016 Oct 27. Bioconjug Chem. 2017. PMID: 27737540 Free PMC article.
-
Intrinsically disordered proteins in synaptic vesicle trafficking and release.J Biol Chem. 2019 Mar 8;294(10):3325-3342. doi: 10.1074/jbc.REV118.006493. Epub 2019 Jan 30. J Biol Chem. 2019. PMID: 30700558 Free PMC article. Review.
-
The origin and impact of bound water around intrinsically disordered proteins.Biophys J. 2022 Feb 15;121(4):540-551. doi: 10.1016/j.bpj.2022.01.011. Epub 2022 Jan 21. Biophys J. 2022. PMID: 35074392 Free PMC article.
-
Revealing the Dynamical Role of Co-solvents in the Coupled Folding and Dimerization of Insulin.J Phys Chem Lett. 2020 Jun 4;11(11):4353-4358. doi: 10.1021/acs.jpclett.0c00982. Epub 2020 May 19. J Phys Chem Lett. 2020. PMID: 32401513 Free PMC article.
-
Disruption of the MBD2-NuRD complex but not MBD3-NuRD induces high level HbF expression in human adult erythroid cells.Haematologica. 2019 Dec;104(12):2361-2371. doi: 10.3324/haematol.2018.210963. Epub 2019 Apr 19. Haematologica. 2019. PMID: 31004025 Free PMC article.
References
-
- Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., and Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 - PubMed
-
- van der Lee R., Buljan M., Lang B., Weatheritt R. J., Daughdrill G. W., Dunker A. K., Fuxreiter M., Gough J., Gsponer J., Jones D. T., Kim P. M., Kriwacki R. W., Oldfield C. J., Pappu R. V., Tompa P., Uversky V. N., Wright P. E., and Babu M. M. (2014) Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 - PMC - PubMed
-
- Jensen M. R., Ruigrok R. W. H., and Blackledge M. (2013) Describing intrinsically disordered proteins at atomic resolution by NMR. Curr. Opin. Struct. Biol. 23, 426–435 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

