Expanding the Range of Protein Function at the Far End of the Order-Structure Continuum
- PMID: 26851282
- PMCID: PMC4807258
- DOI: 10.1074/jbc.R115.692590
Expanding the Range of Protein Function at the Far End of the Order-Structure Continuum
Abstract
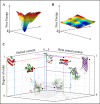
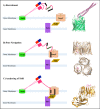
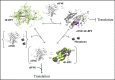
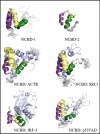
The traditional view of the structure-function paradigm is that a protein's function is inextricably linked to a well defined, three-dimensional structure, which is determined by the protein's primary amino acid sequence. However, it is now accepted that a number of proteins do not adopt a unique tertiary structure in solution and that some degree of disorder is required for many proteins to perform their prescribed functions. In this review, we highlight how a number of protein functions are facilitated by intrinsic disorder and introduce a new protein structure taxonomy that is based on quantifiable metrics of a protein's disorder.
Keywords: biophysics; conformational change; intrinsically disordered protein; protein folding; protein structure.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- McCammon J. A., Gelin B. R., and Karplus M. (1977) Dynamics of folded proteins. Nature 267, 585–590 - PubMed
-
- Wright P. E., and Dyson H. J. (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331 - PubMed
-
- Marsh J. A., Teichmann S. A., and Forman-Kay J. D. (2012) Probing the diverse landscape of protein flexibility and binding. Curr. Opin Struct. Biol. 22, 643–650 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

