pMINERVA: A donor-acceptor system for the in vivo recombineering of scFv into IgG molecules
- PMID: 26851519
- PMCID: PMC4792691
- DOI: 10.1016/j.jim.2016.02.003
pMINERVA: A donor-acceptor system for the in vivo recombineering of scFv into IgG molecules
Abstract
Phage display is the most widely used method for selecting binding molecules from recombinant antibody libraries. However, validation of the phage antibodies often requires early production of the cognate full-length immunoglobulin G (IgG). The conversion of phage library outputs to a full immunoglobulin via standard subcloning is time-consuming and limits the number of clones that can be evaluated. We have developed a novel system to convert scFvs from a phage display vector directly into IgGs without any in vitro subcloning steps. This new vector system, named pMINERVA, makes clever use of site-specific bacteriophage integrases that are expressed in Escherichia coli and intron splicing that occurs within mammalian cells. Using this system, a phage display vector contains both bacterial and mammalian regulatory regions that support antibody expression in E. coli and mammalian cells. A single-chain variable fragment (scFv) antibody is expressed on the surface of bacteriophage M13 as a genetic fusion to the gpIII coat protein. The scFv is converted to an IgG that can be expressed in mammalian cells by transducing a second E. coli strain. In that strain, the phiC31 recombinase fuses the heavy chain constant domain from an acceptor plasmid to the heavy chain variable domain and introduces controlling elements upstream of the light chain variable domain. Splicing in mammalian cells removes a synthetic intron containing the M13 gpIII gene to produce the fusion of the light chain variable domain to the constant domain. We show that phage displaying a scFv and recombinant IgGs generated using this system are expressed at wild-type levels and retain normal function. Use of the pMINERVA completely eliminates the labor-intensive subcloning and DNA sequence confirmation steps currently needed to convert a scFv into a functional IgG Ab.
Keywords: IgG; Recombineering; ScFv; Splicing.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
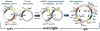



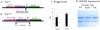
References
-
- Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10:712. - PubMed
-
- Bradbury A, Pluckthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. - PubMed
-
- Bradbury AM, Pluckthun A. Antibodies: validate recombinants once. Nature. 2015;520:295. - PubMed
-
- Taussig MJ, Schmidt R, Cook EA, Stoevesandt O. Development of proteome-wide binding reagents for research and diagnostics. Proteomics Clin Appl. 2013;7:756–766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

