Rapid addition of unlabeled silent solubility tags to proteins using a new substrate-fused sortase reagent
- PMID: 26852413
- PMCID: PMC5110246
- DOI: 10.1007/s10858-016-0019-z
Rapid addition of unlabeled silent solubility tags to proteins using a new substrate-fused sortase reagent
Abstract
Many proteins can't be studied using solution NMR methods because they have limited solubility. To overcome this problem, recalcitrant proteins can be fused to a more soluble protein that functions as a solubility tag. However, signals arising from the solubility tag hinder data analysis because they increase spectral complexity. We report a new method to rapidly and efficiently add a non-isotopically labeled Small Ubiquitin-like Modifier protein (SUMO) solubility tag to an isotopically labeled protein. The method makes use of a newly developed SUMO-Sortase tagging reagent in which SUMO and the Sortase A (SrtA) enzyme are present within the same polypeptide. The SUMO-Sortase reagent rapidly attaches SUMO to any protein that contains the sequence LPXTG at its C-terminus. It modifies proteins at least 15-times faster than previously described approaches, and does not require active dialysis or centrifugation during the reaction to increase product yields. In addition, silently tagged proteins are readily purified using the well-established SUMO expression and purification system. The utility of the SUMO-Sortase tagging reagent is demonstrated using PhoP and green fluorescent proteins, which are ~90% modified with SUMO at room temperature within four hours. SrtA is widely used as a tool to construct bioconjugates. Significant rate enhancements in these procedures may also be achieved by fusing the sortase enzyme to its nucleophile substrate.
Keywords: Protein ligation; SUMO; Silent solubility tag; Sortase.
Figures


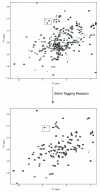
References
-
- Cavanagh J, Fairbrother WJ, Palmer AG, et al. Protein NMR Spectroscopy: Principles and Practice. Academic Press; 2010.
-
- Delaglio F, Grzesiek S, Vuister GW, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

