Colorectal cancer characterization and therapeutic target prediction based on microRNA expression profile
- PMID: 26852921
- PMCID: PMC4745004
- DOI: 10.1038/srep20616
Colorectal cancer characterization and therapeutic target prediction based on microRNA expression profile
Abstract
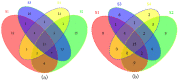
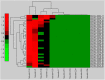
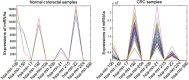
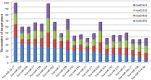
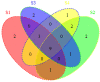
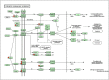
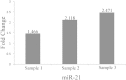
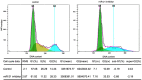
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers and a major cause of cancer death. However, the molecular mechanisms underlying CRC initiation, growth and metastasis are poorly understood. In this study, based on our previous work for comprehensively analyzing miRNA sequencing data, we examined a series of colorectal cancer microRNAs expression profiles data. Results show that all these CRC samples share the same four pathways including TGF-beta signaling pathway, which is important in colorectal carcinogenesis. Twenty-one microRNAs that evolved in the four overlapped pathways were then discovered. Further analysis selected miR-21 as an important regulator for CRC through TGF-beta pathways. This study develops methods for discovering tumor specific miRNA cluster as biomarker and for screening new cancer therapy targets based on miRNA sequencing.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

