Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity
- PMID: 26853474
- PMCID: PMC4752873
- DOI: 10.1016/j.cell.2015.12.053
Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity
Abstract
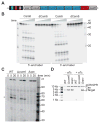
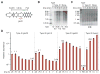
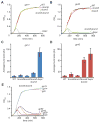
Type III-A CRISPR-Cas systems defend prokaryotes against viral infection using CRISPR RNA (crRNA)-guided nucleases that perform co-transcriptional cleavage of the viral target DNA and its transcripts. Whereas DNA cleavage is essential for immunity, the function of RNA targeting is unknown. Here, we show that transcription-dependent targeting results in a sharp increase of viral genomes in the host cell when the target is located in a late-expressed phage gene. In this targeting condition, mutations in the active sites of the type III-A RNases Csm3 and Csm6 lead to the accumulation of the target phage mRNA and abrogate immunity. Csm6 is also required to provide defense in the presence of mutated phage targets, when DNA cleavage efficiency is reduced. Our results show that the degradation of phage transcripts by CRISPR-associated RNases ensures robust immunity in situations that lead to a slow clearance of the target DNA.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

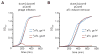

Comment in
-
CRISPR-Cas Gatekeeper: Slow on the Uptake but Gets the Job Done.Cell Host Microbe. 2016 Feb 10;19(2):135-7. doi: 10.1016/j.chom.2016.01.015. Cell Host Microbe. 2016. PMID: 26867170
-
Bacterial physiology: It's never too late for CRISPR RNases.Nat Rev Microbiol. 2016 Apr;14(4):192. doi: 10.1038/nrmicro.2016.28. Epub 2016 Feb 22. Nat Rev Microbiol. 2016. PMID: 26898366 No abstract available.
References
-
- Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006;62:1035–1047. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgartner M, Bonneau F, Mann M, Conti E. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

