H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA
- PMID: 26853553
- PMCID: PMC4745008
- DOI: 10.1038/srep20121
H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA
Abstract
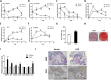
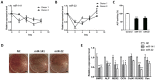
Bone homeostasis is tightly orchestrated and maintained by the balance between osteoblasts and osteoclasts. Recent studies have greatly expanded our understanding of the molecular mechanisms of cellular differentiation. However, the functional roles of non-coding RNAs particularly lncRNAs in remodeling bone architecture remain elusive. In our study, lncRNA H19 was found to be upregulated during osteogenesis in hMSCs. Stable expression of H19 significantly accelerated in vivo and in vitro osteoblast differentiation. Meanwhile, by using bioinformatic investigations and RIP assays combined with luciferase reporter assays, we demonstrated that H19 functioned as an miRNA sponge for miR-141 and miR-22, both of which were negative regulators of osteogenesis and Wnt/β-catenin pathway. Further investigations revealed that H19 antagonized the functions of these two miRNAs and led to de-repression of their shared target gene β-catenin, which eventually activated Wnt/β-catenin pathway and hence potentiated osteogenesis. In addition, we also identified a novel regulatory feedback loop between H19 and its encoded miR-675-5p. And miR-675-5p was found to directly target H19 and counteracted osteoblast differentiation. To sum up, these observations indicate that the lncRNA H19 modulates Wnt/β-catenin pathway by acting as a competing endogenous RNA, which may shed light on the functional role of lncRNAs in coordinating osteogenesis.
Figures





Similar articles
-
MiR-214 inhibits human mesenchymal stem cells differentiating into osteoblasts through targeting β-catenin.Eur Rev Med Pharmacol Sci. 2017 Nov;21(21):4777-4783. Eur Rev Med Pharmacol Sci. 2017. PMID: 29164587
-
H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/β-catenin in ox-LDL -stimulated vascular smooth muscle cells.J Biomed Sci. 2018 Feb 7;25(1):11. doi: 10.1186/s12929-018-0418-4. J Biomed Sci. 2018. PMID: 29415742 Free PMC article.
-
Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-β1/Smad3/HDAC Signaling Pathway by Deriving miR-675.Stem Cells. 2015 Dec;33(12):3481-92. doi: 10.1002/stem.2225. Epub 2015 Oct 23. Stem Cells. 2015. PMID: 26417995
-
[Regulation of microRNA in osteoblast differentiation and its clinical significance].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2012 Jun;26(6):755-9. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2012. PMID: 22792778 Review. Chinese.
-
[Research progress on the effect of long non-coding RNA H19 on osteogenic differentiation and bone diseases].Sheng Li Xue Bao. 2018 Oct 25;70(5):531-538. Sheng Li Xue Bao. 2018. PMID: 30377692 Review. Chinese.
Cited by
-
LncRNA H19 Overexpression Activates Wnt Signaling to Maintain the Hair Follicle Regeneration Potential of Dermal Papilla Cells.Front Genet. 2020 Aug 4;11:694. doi: 10.3389/fgene.2020.00694. eCollection 2020. Front Genet. 2020. PMID: 32849769 Free PMC article.
-
LncRNA-PCAT1 targeting miR-145-5p promotes TLR4-associated osteogenic differentiation of adipose-derived stem cells.J Cell Mol Med. 2018 Dec;22(12):6134-6147. doi: 10.1111/jcmm.13892. Epub 2018 Oct 19. J Cell Mol Med. 2018. PMID: 30338912 Free PMC article.
-
The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-Catenin pathway.Onco Targets Ther. 2018 Jan 11;11:313-321. doi: 10.2147/OTT.S154339. eCollection 2018. Onco Targets Ther. 2018. Retraction in: Onco Targets Ther. 2022 Apr 07;15:329-330. doi: 10.2147/OTT.S369352. PMID: 29391808 Free PMC article. Retracted.
-
LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells.Front Genet. 2020 Mar 31;11:277. doi: 10.3389/fgene.2020.00277. eCollection 2020. Front Genet. 2020. PMID: 32296461 Free PMC article. Review.
-
LncRNA H19 mediates BMP9-induced angiogenesis in mesenchymal stem cells by promoting the p53-Notch1 angiogenic signaling axis.Genes Dis. 2022 May 10;10(3):1040-1054. doi: 10.1016/j.gendis.2022.04.013. eCollection 2023 May. Genes Dis. 2022. PMID: 37396541 Free PMC article.
References
-
- Phan T. C. A., Xu J. & Zheng M. H. Interaction between osteoblast and osteoclast: impact in bone disease. Histol Histopathol 19, 1325–1344 (2004). - PubMed
-
- Pittenger M. F. et al. Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 (1999). - PubMed
-
- Zhang W. X., Yang N. L. & Shi X. M. Regulation of mesenchymal stem cell osteogenic differentiation by glucocorticoid-induced leucine zipper (GILZ). J Biol Chem 283, 4723–4729 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

