MicroRNAs activate gene transcription epigenetically as an enhancer trigger
- PMID: 26853707
- PMCID: PMC5711461
- DOI: 10.1080/15476286.2015.1112487
MicroRNAs activate gene transcription epigenetically as an enhancer trigger
Abstract
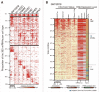
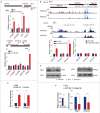
MicroRNAs (miRNAs) are small non-coding RNAs that function as negative gene expression regulators. Emerging evidence shows that, except for function in the cytoplasm, miRNAs are also present in the nucleus. However, the functional significance of nuclear miRNAs remains largely undetermined. By screening miRNA database, we have identified a subset of miRNA that functions as enhancer regulators. Here, we found a set of miRNAs show gene-activation function. We focused on miR-24-1 and found that this miRNA unconventionally activates gene transcription by targeting enhancers. Consistently, the activation was completely abolished when the enhancer sequence was deleted by TALEN. Furthermore, we found that miR-24-1 activates enhancer RNA (eRNA) expression, alters histone modification, and increases the enrichment of p300 and RNA Pol II at the enhancer locus. Our results demonstrate a novel mechanism of miRNA as an enhancer trigger.
Keywords: Chromatin modification; enhancer; miR-24; microRNA; nucleus; transcriptional activation.
Figures



Similar articles
-
miRNA-Mediated RNAa by Targeting Enhancers.Adv Exp Med Biol. 2017;983:113-125. doi: 10.1007/978-981-10-4310-9_8. Adv Exp Med Biol. 2017. PMID: 28639195
-
A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription.Mol Cell. 2017 Jul 20;67(2):308-321.e6. doi: 10.1016/j.molcel.2017.06.028. Mol Cell. 2017. PMID: 28732206 Free PMC article.
-
The histone variant H2A.Z is an important regulator of enhancer activity.Nucleic Acids Res. 2015 Nov 16;43(20):9742-56. doi: 10.1093/nar/gkv825. Epub 2015 Aug 28. Nucleic Acids Res. 2015. PMID: 26319018 Free PMC article.
-
Enhancer RNA: biogenesis, function, and regulation.Essays Biochem. 2020 Dec 7;64(6):883-894. doi: 10.1042/EBC20200014. Essays Biochem. 2020. PMID: 33034351 Review.
-
Enhancer RNAs: transcriptional regulators and workmates of NamiRNAs in myogenesis.Cell Mol Biol Lett. 2021 Feb 10;26(1):4. doi: 10.1186/s11658-021-00248-x. Cell Mol Biol Lett. 2021. PMID: 33568070 Free PMC article. Review.
Cited by
-
Improvement of hyperlipidemia by aerobic exercise in mice through a regulatory effect of miR-21a-5p on its target genes.Sci Rep. 2021 Jun 7;11(1):11966. doi: 10.1038/s41598-021-91583-8. Sci Rep. 2021. PMID: 34099844 Free PMC article.
-
Identification of miRNA-eQTLs in maize mature leaf by GWAS.BMC Genomics. 2020 Oct 6;21(1):689. doi: 10.1186/s12864-020-07073-0. BMC Genomics. 2020. PMID: 33023467 Free PMC article.
-
NamiRNA-mediated high expression of KNSTRN correlates with poor prognosis and immune infiltration in hepatocellular carcinoma.Contemp Oncol (Pozn). 2023;27(3):163-175. doi: 10.5114/wo.2023.133507. Epub 2023 Dec 6. Contemp Oncol (Pozn). 2023. PMID: 38239867 Free PMC article.
-
Molecular mechanisms of transgenerational epigenetic inheritance.Nat Rev Genet. 2022 Jun;23(6):325-341. doi: 10.1038/s41576-021-00438-5. Epub 2022 Jan 4. Nat Rev Genet. 2022. PMID: 34983971 Review.
-
Multi-Omics Reveal the Immunological Role and the Theragnostic Value of miR-216a/GDF15 Axis in Human Colon Adenocarcinoma.Int J Mol Sci. 2021 Dec 20;22(24):13636. doi: 10.3390/ijms222413636. Int J Mol Sci. 2021. PMID: 34948431 Free PMC article.
References
-
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010; 79:351-79; PMID:20533884; https://doi.org/10.1146/annurev-biochem-060308-103103 - DOI - PubMed
-
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012; 13:271-82; PMID:22411466; https://doi.org/10.1038/nrg3162 - DOI - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; https://doi.org/10.1016/S0092-8674(04)00045-5 - DOI - PubMed
-
- Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 2013; 41:10086-109; PMID:23999091; https://doi.org/10.1093/nar/gkt777 - DOI - PMC - PubMed
-
- Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010; 116:5637-49; PMID:20737563; https://doi.org/10.1002/cncr.25488 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
