Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory
- PMID: 26853856
- PMCID: PMC4779431
- DOI: 10.1016/j.stem.2016.01.019
Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory
Abstract
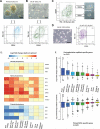
Human embryonic stem cells (hESCs) typically exhibit "primed" pluripotency, analogous to stem cells derived from the mouse post-implantation epiblast. This has led to a search for growth conditions that support self-renewal of hESCs akin to hypomethylated naive epiblast cells in human pre-implantation embryos. We have discovered that reverting primed hESCs to a hypomethylated naive state or deriving a new hESC line under naive conditions results in the establishment of Stage Specific Embryonic Antigen 4 (SSEA4)-negative hESC lines with a transcriptional program resembling the human pre-implantation epiblast. In contrast, we discovered that the methylome of naive hESCs in vitro is distinct from that of the human epiblast in vivo with loss of DNA methylation at primary imprints and a lost "memory" of the methylation state of the human oocyte. This failure to recover the naive epiblast methylation landscape appears to be a consistent feature of self-renewing hypomethylated naive hESCs in vitro.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

References
-
- Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. - PubMed
-
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
