Expression of the MOZ-TIF2 oncoprotein in mice represses senescence
- PMID: 26854485
- PMCID: PMC4819447
- DOI: 10.1016/j.exphem.2015.12.006
Expression of the MOZ-TIF2 oncoprotein in mice represses senescence
Abstract
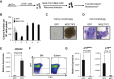
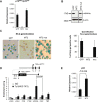
The MOZ-TIF2 translocation, which fuses monocytic leukemia zinc finger protein (MOZ) histone acetyltransferase (HAT) with the nuclear co-activator TIF2, is associated with the development of acute myeloid leukemia. We recently found that in the absence of MOZ HAT activity, p16(INK4a) transcriptional levels are significantly increased, triggering an early entrance into replicative senescence. Because oncogenic fusion proteins must bypass cellular safeguard mechanisms, such as senescence and apoptosis, to induce leukemia, we hypothesized that this repressive activity of MOZ over p16(INK4a) transcription could be preserved, or even reinforced, in MOZ leukemogenic fusion proteins, such as MOZ-TIF2. We describe here that, indeed, MOZ-TIF2 silences expression of the CDKN2A locus (p16(INK4a) and p19(ARF)), inhibits the triggering of senescence and enhances proliferation, providing conditions favorable to the development of leukemia. Furthermore, we describe that abolishing the MOZ HAT activity of the fusion protein leads to a significant increase in expression of the CDKN2A locus and the number of hematopoietic progenitors undergoing senescence. Finally, we report that inhibition of senescence by MOZ-TIF2 is associated with increased apoptosis, suggesting a role for the fusion protein in p53 apoptosis-versus-senescence balance. Our results underscore the importance of the HAT activity of MOZ, preserved in the fusion protein, for repression of the CDKN2A locus transcription and the subsequent block of senescence, a necessary step for the survival of leukemic cells.
Copyright © 2016 ISEH - International Society for Experimental Hematology. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
MOZ-mediated repression of p16(INK) (4) (a) is critical for the self-renewal of neural and hematopoietic stem cells.Stem Cells. 2014 Jun;32(6):1591-601. doi: 10.1002/stem.1606. Stem Cells. 2014. PMID: 24307508 Free PMC article.
-
MOZ-TIF2 repression of nuclear receptor-mediated transcription requires multiple domains in MOZ and in the CID domain of TIF2.Mol Cancer. 2007 Aug 13;6:51. doi: 10.1186/1476-4598-6-51. Mol Cancer. 2007. PMID: 17697320 Free PMC article.
-
A MOZ-TIF2 leukemia mouse model displays KAT6-dependent H3K23 propionylation and overexpression of a set of active developmental genes.Proc Natl Acad Sci U S A. 2024 Jun 25;121(26):e2405905121. doi: 10.1073/pnas.2405905121. Epub 2024 Jun 18. Proc Natl Acad Sci U S A. 2024. PMID: 38889153 Free PMC article.
-
MOZ fusion proteins in acute myeloid leukaemia.Biochem Soc Symp. 2006;(73):23-39. doi: 10.1042/bss0730023. Biochem Soc Symp. 2006. PMID: 16626284 Review.
-
Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis.Cancer Sci. 2008 Aug;99(8):1523-7. doi: 10.1111/j.1349-7006.2008.00865.x. Cancer Sci. 2008. PMID: 18754862 Free PMC article. Review.
Cited by
-
The MOZ-BRPF1 acetyltransferase complex in epigenetic crosstalk linked to gene regulation, development, and human diseases.Front Cell Dev Biol. 2023 Jan 11;10:1115903. doi: 10.3389/fcell.2022.1115903. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36712963 Free PMC article. Review.
-
The Biological Significance of Targeting Acetylation-Mediated Gene Regulation for Designing New Mechanistic Tools and Potential Therapeutics.Biomolecules. 2021 Mar 18;11(3):455. doi: 10.3390/biom11030455. Biomolecules. 2021. PMID: 33803759 Free PMC article. Review.
-
Emerging Roles of GLI-Similar Krüppel-like Zinc Finger Transcription Factors in Leukemia and Other Cancers.Trends Cancer. 2019 Sep;5(9):547-557. doi: 10.1016/j.trecan.2019.07.005. Epub 2019 Aug 20. Trends Cancer. 2019. PMID: 31474360 Free PMC article. Review.
-
The many lives of KATs - detectors, integrators and modulators of the cellular environment.Nat Rev Genet. 2019 Jan;20(1):7-23. doi: 10.1038/s41576-018-0072-4. Nat Rev Genet. 2019. PMID: 30390049 Review.
-
Chromatin accessibility promotes hematopoietic and leukemia stem cell activity.Nat Commun. 2020 Mar 16;11(1):1406. doi: 10.1038/s41467-020-15221-z. Nat Commun. 2020. PMID: 32179749 Free PMC article.
References
-
- Avvakumov N., Côté J. Functions of myst family histone acetyltransferases and their link to disease. Subcell Biochem. 2007;41:295–317. - PubMed
-
- Borrow J., Stanton V.P., Jr., Andresen J.M. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

