Increased retinal mtDNA damage in the CFH variant associated with age-related macular degeneration
- PMID: 26854823
- PMCID: PMC4842097
- DOI: 10.1016/j.exer.2016.01.018
Increased retinal mtDNA damage in the CFH variant associated with age-related macular degeneration
Abstract
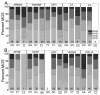
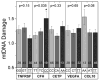
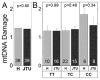
Age-related macular degeneration (AMD) is a major cause of blindness among the elderly in the developed world. Genetic analysis of AMD has identified 34 high-risk loci associated with AMD. The genes at these high risk loci belong to diverse biological pathways, suggesting different mechanisms leading to AMD pathogenesis. Thus, therapies targeting a single pathway for all AMD patients will likely not be universally effective. Recent evidence suggests defects in mitochondria (mt) of the retinal pigment epithelium (RPE) may constitute a key pathogenic event in some AMD patients. The purpose of this study is to determine if individuals with a specific genetic background have a greater propensity for mtDNA damage. We used human eyebank tissues from 76 donors with AMD and 42 age-matched controls to determine the extent of mtDNA damage in the RPE that was harvested from the macula using a long extension polymerase chain reaction assay. Genotype analyses were performed for ten common AMD-associated nuclear risk alleles (ARMS2, TNFRSF10A, CFH, C2, C3, APOE, CETP, LIPC, VEGF and COL10A1) and mtDNA haplogroups. Sufficient samples were available for genotype association with mtDNA damage for TNFRSF10A, CFH, CETP, VEGFA, and COL10A1. Our results show that AMD donors carrying the high risk allele for CFH (C) had significantly more mtDNA damage compared with donors having the wild-type genetic profile. The data from an additional 39 donors (12 controls and 27 AMD) genotyped for CFH alleles further supported these findings. Taken together, these studies provide the rationale for a more personalized approach for treating AMD by uncovering a significant correlation between the CFH high risk allele and accelerated mtDNA damage. Patients harboring this genetic risk factor may benefit from therapies that stabilize and protect the mt in the RPE.
Keywords: Age-related macular degeneration; Complement factor H; Eyebank tissue; Haplogroups; Inflammation; Mitochondria; mtDNA.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures


References
-
- Atilano SR, Malik D, Chwa M, Cáceres-Del-Carpio J, Nesburn AB, Boyer DS, Kuppermann BD, Jazwinski SM, Miceli MV, Wallace DC, et al. Mitochondrial DNA variants can mediate methylation status of inflammation, angiogenesis and signaling genes. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv173. - DOI - PMC - PubMed
-
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. PNAS. 2010;107:7401–7406. - PMC - PubMed
-
- Chew EY, Klein ML, Clemons TE, Agrón E, Ratnapryiya R, Edwards AO, Fritsche LG, Swaroop A, Abecasis GR Age-Related Eye Disease Study Research Group. No clinically significant association between CFH and ARMS2 genotypes and response to nutritional supplements: AREDS report number 38. Ophthalmology. 2014;121:2173–2180. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

