Selectivity profiling of BCRP versus P-gp inhibition: from automated collection of polypharmacology data to multi-label learning
- PMID: 26855674
- PMCID: PMC4743411
- DOI: 10.1186/s13321-016-0121-y
Selectivity profiling of BCRP versus P-gp inhibition: from automated collection of polypharmacology data to multi-label learning
Abstract
Background: The human ATP binding cassette transporters Breast Cancer Resistance Protein (BCRP) and Multidrug Resistance Protein 1 (P-gp) are co-expressed in many tissues and barriers, especially at the blood-brain barrier and at the hepatocyte canalicular membrane. Understanding their interplay in affecting the pharmacokinetics of drugs is of prime interest. In silico tools to predict inhibition and substrate profiles towards BCRP and P-gp might serve as early filters in the drug discovery and development process. However, to build such models, pharmacological data must be collected for both targets, which is a tedious task, often involving manual and poorly reproducible steps.
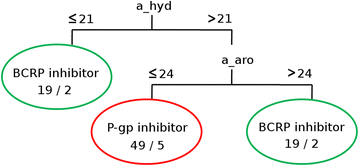
Results: Compounds with inhibitory activity measured against BCRP and/or P-gp were retrieved by combining Open Data and manually curated data from literature using a KNIME workflow. After determination of compound overlap, machine learning approaches were used to establish multi-label classification models for BCRP/P-gp. Different ways of addressing multi-label problems are explored and compared: label-powerset, binary relevance and classifiers chain. Label-powerset revealed important molecular features for selective or polyspecific inhibitory activity. In our dataset, only two descriptors (the numbers of hydrophobic and aromatic atoms) were sufficient to separate selective BCRP inhibitors from selective P-gp inhibitors. Also, dual inhibitors share properties with both groups of selective inhibitors. Binary relevance and classifiers chain allow improving the predictivity of the models.
Conclusions: The KNIME workflow proved a useful tool to merge data from diverse sources. It could be used for building multi-label datasets of any set of pharmacological targets for which there is data available either in the open domain or in-house. By applying various multi-label learning algorithms, important molecular features driving transporter selectivity could be retrieved. Finally, using the dataset with missing annotations, predictive models can be derived in cases where no accurate dense dataset is available (not enough data overlap or no well balanced class distribution).Graphical abstract.
Keywords: BCRP; Binary relevance; Classifiers chain; KNIME; Multi-label classification; Open Data; Open PHACTS; P-glycoprotein; Polyspecific inhibition; Selective inhibition.
Figures




Similar articles
-
In vitro assessment of the interactions of dopamine β-hydroxylase inhibitors with human P-glycoprotein and Breast Cancer Resistance Protein.Eur J Pharm Sci. 2018 May 30;117:35-40. doi: 10.1016/j.ejps.2018.02.006. Epub 2018 Feb 8. Eur J Pharm Sci. 2018. PMID: 29428540
-
Subtle Structural Differences Trigger Inhibitory Activity of Propafenone Analogues at the Two Polyspecific ABC Transporters: P-Glycoprotein (P-gp) and Breast Cancer Resistance Protein (BCRP).ChemMedChem. 2016 Jun 20;11(12):1380-94. doi: 10.1002/cmdc.201500592. Epub 2016 Mar 10. ChemMedChem. 2016. PMID: 26970257 Free PMC article.
-
Metabolomic Profiling and Drug Interaction Characterization Reveal Riboflavin As a Breast Cancer Resistance Protein-Specific Endogenous Biomarker That Demonstrates Prediction of Transporter Activity In Vivo.Drug Metab Dispos. 2023 Jul;51(7):851-861. doi: 10.1124/dmd.123.001284. Epub 2023 Apr 13. Drug Metab Dispos. 2023. PMID: 37055191
-
Clinical Relevance of Hepatic and Renal P-gp/BCRP Inhibition of Drugs: An International Transporter Consortium Perspective.Clin Pharmacol Ther. 2022 Sep;112(3):573-592. doi: 10.1002/cpt.2670. Epub 2022 Jun 22. Clin Pharmacol Ther. 2022. PMID: 35612761 Free PMC article. Review.
-
Recent advances in the search of BCRP- and dual P-gp/BCRP-based multidrug resistance modulators.Cancer Drug Resist. 2019 Sep 19;2(3):710-743. doi: 10.20517/cdr.2019.31. eCollection 2019. Cancer Drug Resist. 2019. PMID: 35582565 Free PMC article. Review.
Cited by
-
Ligand- and Structure-based Approaches for Transmembrane Transporter Modeling.Curr Drug Res Rev. 2024;16(2):81-93. doi: 10.2174/2589977515666230508123041. Curr Drug Res Rev. 2024. PMID: 37157206 Free PMC article. Review.
-
ADMET evaluation in drug discovery. 20. Prediction of breast cancer resistance protein inhibition through machine learning.J Cheminform. 2020 Mar 5;12(1):16. doi: 10.1186/s13321-020-00421-y. J Cheminform. 2020. PMID: 33430990 Free PMC article.
-
How Open Data Shapes In Silico Transporter Modeling.Molecules. 2017 Mar 7;22(3):422. doi: 10.3390/molecules22030422. Molecules. 2017. PMID: 28272367 Free PMC article. Review.
-
A model for identifying potentially inappropriate medication used in older people with dementia: a machine learning study.Int J Clin Pharm. 2024 Aug;46(4):937-946. doi: 10.1007/s11096-024-01730-0. Epub 2024 Jul 9. Int J Clin Pharm. 2024. PMID: 38980590 Free PMC article.
-
Current Advances in Studying Clinically Relevant Transporters of the Solute Carrier (SLC) Family by Connecting Computational Modeling and Data Science.Comput Struct Biotechnol J. 2019 Mar 8;17:390-405. doi: 10.1016/j.csbj.2019.03.002. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 30976382 Free PMC article. Review.
References
-
- Gray AJG, Groth P, Loizou A, Askjaer S, Brenninkmeijer C, Burger K, Chichester C, Evelo CT, Goble C, Harland L, Pettifer S, Thompson M, Waagmeester A, Williams AJ. Applying linked data approaches to pharmacology: architectural decisions and implementation. Semant Web. 2014;5:101–113. doi: 10.5121/ijwest.2014.5407. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

