Cell type-specific properties and environment shape tissue specificity of cancer genes
- PMID: 26856619
- PMCID: PMC4746590
- DOI: 10.1038/srep20707
Cell type-specific properties and environment shape tissue specificity of cancer genes
Abstract
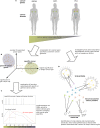
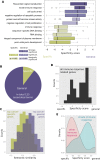
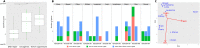
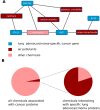
One of the biggest mysteries in cancer research remains why mutations in certain genes cause cancer only at specific sites in the human body. The poor correlation between the expression level of a cancer gene and the tissues in which it causes malignant transformations raises the question of which factors determine the tissue-specific effects of a mutation. Here, we explore why some cancer genes are associated only with few different cancer types (i.e., are specific), while others are found mutated in a large number of different types of cancer (i.e., are general). We do so by contrasting cellular functions of specific-cancer genes with those of general ones to identify properties that determine where in the body a gene mutation is causing malignant transformations. We identified different groups of cancer genes that did not behave as expected (i.e., DNA repair genes being tissue specific, immune response genes showing a bimodal specificity function or strong association of generally expressed genes to particular cancers). Analysis of these three groups demonstrates the importance of environmental impact for understanding why certain cancer genes are only involved in the development of some cancer types but are rarely found mutated in other types of cancer.
Figures


Similar articles
-
Dissecting the molecular pathways of (testicular) germ cell tumour pathogenesis; from initiation to treatment-resistance.Int J Androl. 2011 Aug;34(4 Pt 2):e234-51. doi: 10.1111/j.1365-2605.2011.01157.x. Epub 2011 May 12. Int J Androl. 2011. PMID: 21564133 Review.
-
Germ cell tumors of the gonads: a selective review emphasizing problems in differential diagnosis, newly appreciated, and controversial issues.Mod Pathol. 2005 Feb;18 Suppl 2:S61-79. doi: 10.1038/modpathol.3800310. Mod Pathol. 2005. PMID: 15761467 Review.
-
Role of stem cell proteins and microRNAs in embryogenesis and germ cell cancer.Int J Dev Biol. 2013;57(2-4):319-32. doi: 10.1387/ijdb.130020re. Int J Dev Biol. 2013. PMID: 23784843 Review.
-
Mutagenesis and embryonal carcinogenesis.Natl Cancer Inst Monogr. 1979 May;(51):19-24. Natl Cancer Inst Monogr. 1979. PMID: 225668
-
Molecular biology of testicular germ cell tumors: unique features awaiting clinical application.Crit Rev Oncol Hematol. 2014 Mar;89(3):366-85. doi: 10.1016/j.critrevonc.2013.10.001. Epub 2013 Oct 11. Crit Rev Oncol Hematol. 2014. PMID: 24182421 Review.
Cited by
-
The Mutational Landscape of Cancer's Vulnerability to Ionizing Radiation.Clin Cancer Res. 2022 Dec 15;28(24):5343-5358. doi: 10.1158/1078-0432.CCR-22-1914. Clin Cancer Res. 2022. PMID: 36222846 Free PMC article.
-
Dysregulated expression of claudins in cancer.Oncol Lett. 2021 Sep;22(3):641. doi: 10.3892/ol.2021.12902. Epub 2021 Jul 7. Oncol Lett. 2021. PMID: 34386063 Free PMC article. Review.
-
When Do Tumours Develop? Neoplastic Processes Across Different Timescales: Age, Season and Round the Circadian Clock.Evol Appl. 2024 Oct 22;17(10):e70024. doi: 10.1111/eva.70024. eCollection 2024 Oct. Evol Appl. 2024. PMID: 39444444 Free PMC article. Review.
-
Transcriptomic analysis reveals a tissue-specific loss of identity during ageing and cancer.BMC Genomics. 2023 Oct 26;24(1):644. doi: 10.1186/s12864-023-09756-w. BMC Genomics. 2023. PMID: 37884865 Free PMC article.
-
Cancer Type Classification in Liquid Biopsies Based on Sparse Mutational Profiles Enabled through Data Augmentation and Integration.Life (Basel). 2021 Dec 21;12(1):1. doi: 10.3390/life12010001. Life (Basel). 2021. PMID: 35054395 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

