Effects of Low-Dose Amoxicillin on Staphylococcus aureus USA300 Biofilms
- PMID: 26856828
- PMCID: PMC4862544
- DOI: 10.1128/AAC.02070-15
Effects of Low-Dose Amoxicillin on Staphylococcus aureus USA300 Biofilms
Abstract
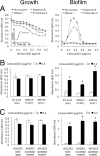
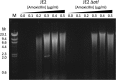
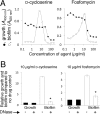
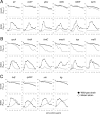
Previous studies showed that sub-MIC levels of β-lactam antibiotics stimulate biofilm formation in most methicillin-resistant Staphylococcus aureus (MRSA) strains. Here, we investigated this process by measuring the effects of sub-MIC amoxicillin on biofilm formation by the epidemic community-associated MRSA strain USA300. We found that sub-MIC amoxicillin increased the ability of USA300 cells to attach to surfaces and form biofilms under both static and flow conditions. We also found that USA300 biofilms cultured in sub-MIC amoxicillin were thicker, contained more pillar and channel structures, and were less porous than biofilms cultured without antibiotic. Biofilm formation in sub-MIC amoxicillin correlated with the production of extracellular DNA (eDNA). However, eDNA released by amoxicillin-induced cell lysis alone was evidently not sufficient to stimulate biofilm. Sub-MIC levels of two other cell wall-active agents with different mechanisms of action-d-cycloserine and fosfomycin-also stimulated eDNA-dependent biofilm, suggesting that biofilm formation may be a mechanistic adaptation to cell wall stress. Screening a USA300 mariner transposon library for mutants deficient in biofilm formation in sub-MIC amoxicillin identified numerous known mediators of S. aureus β-lactam resistance and biofilm formation, as well as novel genes not previously associated with these phenotypes. Our results link cell wall stress and biofilm formation in MRSA and suggest that eDNA-dependent biofilm formation by strain USA300 in low-dose amoxicillin is an inducible phenotype that can be used to identify novel genes impacting MRSA β-lactam resistance and biofilm formation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus.mBio. 2012 Jul 31;3(4):e00198-12. doi: 10.1128/mBio.00198-12. Print 2012. mBio. 2012. PMID: 22851659 Free PMC article.
-
Micrococcal nuclease regulates biofilm formation and dispersal in methicillin-resistant Staphylococcus aureus USA300.mSphere. 2024 May 29;9(5):e0012624. doi: 10.1128/msphere.00126-24. Epub 2024 May 2. mSphere. 2024. PMID: 38695568 Free PMC article.
-
Subinhibitory Concentrations of Mupirocin Stimulate Staphylococcus aureus Biofilm Formation by Upregulating cidA.Antimicrob Agents Chemother. 2020 Feb 21;64(3):e01912-19. doi: 10.1128/AAC.01912-19. Print 2020 Feb 21. Antimicrob Agents Chemother. 2020. PMID: 31932378 Free PMC article.
-
Methicillin resistance and the biofilm phenotype in Staphylococcus aureus.Front Cell Infect Microbiol. 2015 Jan 28;5:1. doi: 10.3389/fcimb.2015.00001. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25674541 Free PMC article. Review.
-
Vision for medicine: Staphylococcus aureus biofilm war and unlocking key's for anti-biofilm drug development.Microb Pathog. 2018 Oct;123:339-347. doi: 10.1016/j.micpath.2018.07.002. Epub 2018 Jul 3. Microb Pathog. 2018. PMID: 30057355 Review.
Cited by
-
Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis.Pathogens. 2021 Oct 21;10(11):1363. doi: 10.3390/pathogens10111363. Pathogens. 2021. PMID: 34832519 Free PMC article.
-
Radezolid Is More Effective Than Linezolid Against Planktonic Cells and Inhibits Enterococcus faecalis Biofilm Formation.Front Microbiol. 2020 Feb 14;11:196. doi: 10.3389/fmicb.2020.00196. eCollection 2020. Front Microbiol. 2020. PMID: 32117185 Free PMC article.
-
Identification of Extracellular DNA-Binding Proteins in the Biofilm Matrix.mBio. 2019 Jun 25;10(3):e01137-19. doi: 10.1128/mBio.01137-19. mBio. 2019. PMID: 31239382 Free PMC article.
-
Microbial Biofilms and Chronic Wounds.Microorganisms. 2017 Mar 7;5(1):9. doi: 10.3390/microorganisms5010009. Microorganisms. 2017. PMID: 28272369 Free PMC article. Review.
-
The role of topical probiotics on wound healing: A review of animal and human studies.Int Wound J. 2020 Dec;17(6):1687-1694. doi: 10.1111/iwj.13451. Epub 2020 Aug 31. Int Wound J. 2020. PMID: 32869480 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

