Specific cell surface labeling of GPCRs using split GFP
- PMID: 26857153
- PMCID: PMC4746647
- DOI: 10.1038/srep20568
Specific cell surface labeling of GPCRs using split GFP
Abstract
Specific cell surface labeling is essential for visualizing the internalization processes of G-protein coupled receptors (GPCRs) and for gaining mechanistic insight of GPCR functions. Here we present a rapid, specific, and versatile labeling scheme for GPCRs at living-cell membrane with the use of a split green fluorescent protein (GFP). Demonstrated with two GPCRs, GPR17 and CysLT2R, we show that two β-stands (β-stands 10 and 11) derived from a superfolder GFP (sfGFP) can be engineered to one of the three extracellular loop of a GPCR. The complementary fragment of sfGFP has nine β-strands (β-stands 1-9) that carries the mature fluorophore, and can be proteolytically derived from the full-length sfGFP. Separately the GFP fragments are non-fluorescent, but become fluorescent upon assembly, thus allowing specific labeling of the target proteins. The two GFP fragments rapidly assemble and the resulting complex is extremely tight under non-denaturing conditions, which allows real-time and quantitative assessment of the internalized GPCRs. We envision that this labeling scheme will be of great use for labeling other membrane proteins in various biological and pharmacological applications.
Figures
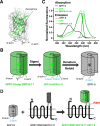
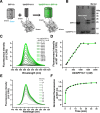
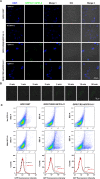
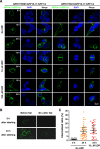
Similar articles
-
Development and Applications of Superfolder and Split Fluorescent Protein Detection Systems in Biology.Int J Mol Sci. 2019 Jul 15;20(14):3479. doi: 10.3390/ijms20143479. Int J Mol Sci. 2019. PMID: 31311175 Free PMC article. Review.
-
Quantitative Measurement of GPCR Endocytosis via Pulse-Chase Covalent Labeling.PLoS One. 2015 May 28;10(5):e0129394. doi: 10.1371/journal.pone.0129394. eCollection 2015. PLoS One. 2015. PMID: 26020647 Free PMC article.
-
Selective covalent labeling of tag-fused GPCR proteins on live cell surface with a synthetic probe for their functional analysis.J Am Chem Soc. 2010 Jul 14;132(27):9301-9. doi: 10.1021/ja910703v. J Am Chem Soc. 2010. PMID: 20568758
-
Quantitative measurement of cell membrane receptor internalization by the nanoluciferase reporter: Using the G protein-coupled receptor RXFP3 as a model.Biochim Biophys Acta. 2015 Feb;1848(2):688-94. doi: 10.1016/j.bbamem.2014.11.026. Epub 2014 Nov 28. Biochim Biophys Acta. 2015. PMID: 25434927
-
Ligand screening system using fusion proteins of G protein-coupled receptors with G protein alpha subunits.Neurochem Int. 2007 Jul-Sep;51(2-4):140-64. doi: 10.1016/j.neuint.2007.06.006. Epub 2007 Jun 19. Neurochem Int. 2007. PMID: 17659814 Review.
Cited by
-
Strategies for Site-Specific Labeling of Receptor Proteins on the Surfaces of Living Cells by Using Genetically Encoded Peptide Tags.Chembiochem. 2021 May 14;22(10):1717-1732. doi: 10.1002/cbic.202000797. Epub 2021 Feb 26. Chembiochem. 2021. PMID: 33428317 Free PMC article. Review.
-
Detecting In-Situ oligomerization of engineered STIM1 proteins by diffraction-limited optical imaging.PLoS One. 2019 Mar 25;14(3):e0213655. doi: 10.1371/journal.pone.0213655. eCollection 2019. PLoS One. 2019. PMID: 30908505 Free PMC article.
-
Development and Applications of Superfolder and Split Fluorescent Protein Detection Systems in Biology.Int J Mol Sci. 2019 Jul 15;20(14):3479. doi: 10.3390/ijms20143479. Int J Mol Sci. 2019. PMID: 31311175 Free PMC article. Review.
-
Hallmarks and Molecular Tools for the Study of Mitophagy in Parkinson's Disease.Cells. 2022 Jul 2;11(13):2097. doi: 10.3390/cells11132097. Cells. 2022. PMID: 35805181 Free PMC article. Review.
-
Quantitative measurement of cell-surface displayed proteins based on split-GFP assembly.Microb Cell Fact. 2024 Apr 12;23(1):108. doi: 10.1186/s12934-024-02386-1. Microb Cell Fact. 2024. PMID: 38609965 Free PMC article.
References
-
- Rask-Andersen M., Almen M. S. & Schioth H. B. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov 10, 579–590 (2011). - PubMed
-
- Fukunaga S., Setoguchi S., Hirasawa A. & Tsujimoto G. Monitoring ligand-mediated internalization of G protein-coupled receptor as a novel pharmacological approach. Life Sci 80, 17–23 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

