Relevance of Sp Binding Site Polymorphism in WWOX for Treatment Outcome in Pancreatic Cancer
- PMID: 26857392
- PMCID: PMC4859408
- DOI: 10.1093/jnci/djv387
Relevance of Sp Binding Site Polymorphism in WWOX for Treatment Outcome in Pancreatic Cancer
Abstract
Background: A genome-wide association study (GWAS) suggested inherited genetic single-nucleotide polymorphisms (SNPs) affecting overall survival (OS) in advanced pancreatic cancer. To identify robust clinical biomarkers, we tested the strongest reported candidate loci in an independent patient cohort, assessed cellular drug sensitivity, and evaluated molecular effects.
Methods: This study comprised 381 patients with histologically verified pancreatic ductal adenocarcinoma treated with gemcitabine-based chemotherapy. The primary outcome was the relationship between germline polymorphisms and OS. Functional assays addressed pharmacological dose-response effects in lymphoblastoid cell lines (LCLs) and pancreatic cancer cell lines (including upon RNAi), gene expression analyses, and allele-specific transcription factor binding. All statistical tests were two-sided.
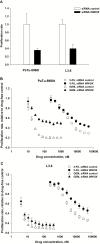
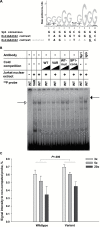
Results: The A allele (26% in Caucasians) at SNP rs11644322 in the putative tumor suppressor gene WWOX conferred worse prognosis. Median OS was 14 months (95% confidence interval [CI] = 12 to 15 months), 13 months (95% CI = 11 to 15 months), and nine months (95% CI = 7 to 12 months) for the GG, GA, and AA genotypes, respectively (P trend < .001 for trend in univariate log-rank assuming a codominant mode of inheritance; advanced disease subgroup P trend < .001). Mean OS was 25 months (95% CI = 21 to 29 months), 19 months (95% CI = 15 to 22 months), and 13 months (95% CI = 10 to 16 months), respectively. This effect held true after adjustment for age, performance status according to Eastern Cooperative Oncology Group classification, TNM, grading, and resection status and was comparable with the strongest established prognostic factors in multivariable analysis. Consistently, reduced responsiveness to gemcitabine, but not 5-fluorouracil, along with lower WWOX expression was demonstrated in LCLs harboring the AA genotype. Likewise, RNAi-mediated WWOX knockdown in pancreatic cancer cells confirmed differential cytostatic drug sensitivity. In electrophoretic mobility shift assays, the A allele exhibited weaker binding of Sp family members Sp1/Sp3.
Conclusions: WWOX rs11644322 represents a major predictive factor in gemcitabine-treated pancreatic cancer. Decreased WWOX expression may interfere with gemcitabine sensitivity, and allele-specific binding at rs11644332 might be a causative molecular mechanism behind the observed clinical associations.
© The Author 2016. Published by Oxford University Press.
Figures



Comment in
-
Genomic Testing for Gemcitabine-Based Treatment of Pancreatic Cancer.J Natl Cancer Inst. 2016 Feb 8;108(5):djv424. doi: 10.1093/jnci/djv424. Print 2016 May. J Natl Cancer Inst. 2016. PMID: 26857393 No abstract available.
Similar articles
-
Genomic Testing for Gemcitabine-Based Treatment of Pancreatic Cancer.J Natl Cancer Inst. 2016 Feb 8;108(5):djv424. doi: 10.1093/jnci/djv424. Print 2016 May. J Natl Cancer Inst. 2016. PMID: 26857393 No abstract available.
-
A randomized, placebo-controlled phase III trial of masitinib plus gemcitabine in the treatment of advanced pancreatic cancer.Ann Oncol. 2015 Jun;26(6):1194-1200. doi: 10.1093/annonc/mdv133. Epub 2015 Apr 9. Ann Oncol. 2015. PMID: 25858497 Free PMC article. Clinical Trial.
-
Single-nucleotide polymorphisms of DNA damage response genes are associated with overall survival in patients with pancreatic cancer.Clin Cancer Res. 2008 Apr 1;14(7):2042-8. doi: 10.1158/1078-0432.CCR-07-1520. Clin Cancer Res. 2008. PMID: 18381943 Free PMC article. Clinical Trial.
-
Mechanisms of drug resistance of pancreatic ductal adenocarcinoma at different levels.Biosci Rep. 2020 Jul 31;40(7):BSR20200401. doi: 10.1042/BSR20200401. Biosci Rep. 2020. PMID: 32677676 Free PMC article. Review.
-
Meta-analysis examining overall survival in patients with pancreatic cancer treated with second-line 5-fluorouracil and oxaliplatin-based therapy after failing first-line gemcitabine-containing therapy: effect of performance status and comparison with other regimens.BMC Cancer. 2020 Jul 8;20(1):633. doi: 10.1186/s12885-020-07110-x. BMC Cancer. 2020. PMID: 32641104 Free PMC article.
Cited by
-
Gene polymorphisms of insulin secretion signaling pathway associated with clopidogrel resistance in Han Chinese population.J Clin Lab Anal. 2021 Nov;35(11):e23970. doi: 10.1002/jcla.23970. Epub 2021 Oct 5. J Clin Lab Anal. 2021. PMID: 34609028 Free PMC article.
-
WWOX Polymorphisms as Predictors of the Biochemical Recurrence of Localized Prostate Cancer after Radical Prostatectomy.Int J Med Sci. 2023 May 21;20(7):969-975. doi: 10.7150/ijms.84364. eCollection 2023. Int J Med Sci. 2023. PMID: 37324196 Free PMC article.
-
Genetic factors affecting patient responses to pancreatic cancer treatment.Ann Gastroenterol. 2016 Oct-Dec;29(4):466-476. doi: 10.20524/aog.2016.0056. Epub 2016 Jun 10. Ann Gastroenterol. 2016. PMID: 27708512 Free PMC article. Review.
-
Single nucleotide polymorphisms and cancer susceptibility.Oncotarget. 2017 Nov 7;8(66):110635-110649. doi: 10.18632/oncotarget.22372. eCollection 2017 Dec 15. Oncotarget. 2017. PMID: 29299175 Free PMC article. Review.
-
Exonic variants in multiple myeloma patients associated with relapsed/ refractory and response to bortezomib regimens.Saudi J Biol Sci. 2022 Jan;29(1):610-614. doi: 10.1016/j.sjbs.2021.09.017. Epub 2021 Sep 16. Saudi J Biol Sci. 2022. PMID: 35002457 Free PMC article.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63 (1):11–30. - PubMed
-
- Burris HA, 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15 (6):2403–2413. - PubMed
-
- Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304 (10):1073–1081. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

