Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+-dependent DNA ligase (LigA)
- PMID: 26857547
- PMCID: PMC4797296
- DOI: 10.1093/nar/gkw049
Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+-dependent DNA ligase (LigA)
Abstract
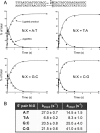
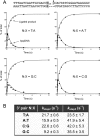
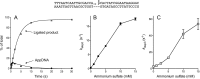
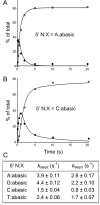
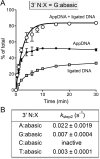
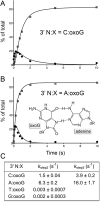
Escherichia coli DNA ligase (EcoLigA) repairs 3'-OH/5'-PO4 nicks in duplex DNA via reaction of LigA with NAD(+) to form a covalent LigA-(lysyl-Nζ)-AMP intermediate (step 1); transfer of AMP to the nick 5'-PO4 to form an AppDNA intermediate (step 2); and attack of the nick 3'-OH on AppDNA to form a 3'-5' phosphodiester (step 3). A distinctive feature of EcoLigA is its stimulation by ammonium ion. Here we used rapid mix-quench methods to analyze the kinetic mechanism of single-turnover nick sealing by EcoLigA-AMP. For substrates with correctly base-paired 3'-OH/5'-PO4 nicks, kstep2 was fast (6.8-27 s(-1)) and similar to kstep3 (8.3-42 s(-1)). Absent ammonium, kstep2 and kstep3 were 48-fold and 16-fold slower, respectively. EcoLigA was exquisitely sensitive to 3'-OH base mispairs and 3' N:abasic lesions, which elicited 1000- to >20000-fold decrements in kstep2. The exception was the non-canonical 3' A:oxoG configuration, which EcoLigA accepted as correctly paired for rapid sealing. These results underscore: (i) how EcoLigA requires proper positioning of the nick 3' nucleoside for catalysis of 5' adenylylation; and (ii) EcoLigA's potential to embed mutations during the repair of oxidative damage. EcoLigA was relatively tolerant of 5'-phosphate base mispairs and 5' N:abasic lesions.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD+-dependent polynucleotide ligases.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2592-2597. doi: 10.1073/pnas.1619220114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223499 Free PMC article.
-
Kinetic mechanism of nick sealing by T4 RNA ligase 2 and effects of 3'-OH base mispairs and damaged base lesions.RNA. 2013 Dec;19(12):1840-7. doi: 10.1261/rna.041731.113. Epub 2013 Oct 24. RNA. 2013. PMID: 24158792 Free PMC article.
-
Analysis of ligation and DNA binding by Escherichia coli DNA ligase (LigA).Biochim Biophys Acta. 2005 May 20;1749(1):113-22. doi: 10.1016/j.bbapap.2005.03.003. Biochim Biophys Acta. 2005. PMID: 15848142
-
DNA and RNA ligases: structural variations and shared mechanisms.Curr Opin Struct Biol. 2008 Feb;18(1):96-105. doi: 10.1016/j.sbi.2007.12.008. Epub 2008 Feb 8. Curr Opin Struct Biol. 2008. PMID: 18262407 Review.
-
DNA ligase: structure, mechanism, and function.Science. 1974 Nov 29;186(4166):790-7. doi: 10.1126/science.186.4166.790. Science. 1974. PMID: 4377758 Review.
Cited by
-
Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.Nucleic Acids Res. 2018 May 4;46(8):4164-4175. doi: 10.1093/nar/gky238. Nucleic Acids Res. 2018. PMID: 29635474 Free PMC article.
-
Excision of the doubly methylated base N4,5-dimethylcytosine from DNA by Escherichia coli Nei and Fpg proteins.Philos Trans R Soc Lond B Biol Sci. 2018 Jun 5;373(1748):20170337. doi: 10.1098/rstb.2017.0337. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29685966 Free PMC article.
-
Alleviation of C⋅C Mismatches in DNA by the Escherichia coli Fpg Protein.Front Microbiol. 2021 Jun 30;12:608839. doi: 10.3389/fmicb.2021.608839. eCollection 2021. Front Microbiol. 2021. PMID: 34276575 Free PMC article.
-
Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD+-dependent polynucleotide ligases.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2592-2597. doi: 10.1073/pnas.1619220114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223499 Free PMC article.
-
Cognate base-pair selectivity of hydrophobic unnatural bases in DNA ligation by T4 DNA ligase.Biopolymers. 2021 Jan;112(1):e23407. doi: 10.1002/bip.23407. Epub 2020 Nov 6. Biopolymers. 2021. PMID: 33156531 Free PMC article.
References
-
- Sriskanda V., Moyer R.W., Shuman S. NAD+-dependent DNA ligase encoded by a eukaryotic virus. J. Biol. Chem. 2001;276:36100–36109. - PubMed
-
- Zhao A, Gray F.C., MacNeill S.A. ATP- and NAD+-dependent DNA ligases share an essential function in the halophilic archaeon Haloferax volcanii. Mol. Microbiol. 2006;59:743–752. - PubMed
-
- Benarroch D., Shuman S. Characterization of mimivirus NAD+-dependent DNA ligase. Virology. 2006;353:133–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

