Light-inducible genetic engineering and control of non-homologous end-joining in industrial eukaryotic microorganisms: LML 3.0 and OFN 1.0
- PMID: 26857594
- PMCID: PMC4746737
- DOI: 10.1038/srep20761
Light-inducible genetic engineering and control of non-homologous end-joining in industrial eukaryotic microorganisms: LML 3.0 and OFN 1.0
Abstract
Filamentous fungi play important roles in the production of plant cell-wall degrading enzymes. In recent years, homologous recombinant technologies have contributed significantly to improved enzymes production and system design of genetically manipulated strains. When introducing multiple gene deletions, we need a robust and convenient way to control selectable marker genes, especially when only a limited number of markers are available in filamentous fungi. Integration after transformation is predominantly nonhomologous in most fungi other than yeast. Fungal strains deficient in the non-homologous end-joining (NHEJ) pathway have limitations associated with gene function analyses despite they are excellent recipient strains for gene targets. We describe strategies and methods to address these challenges above and leverage the power of resilient NHEJ deficiency strains. We have established a foolproof light-inducible platform for one-step unmarked genetic modification in industrial eukaryotic microorganisms designated as 'LML 3.0', and an on-off control protocol of NHEJ pathway called 'OFN 1.0', using a synthetic light-switchable transactivation to control Cre recombinase-based excision and inversion. The methods provide a one-step strategy to sequentially modify genes without introducing selectable markers and NHEJ-deficiency. The strategies can be used to manipulate many biological processes in a wide range of eukaryotic cells.
Figures
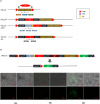


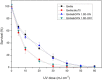

References
-
- Kubicek C. P., Fungi and Lignocellulose Biomass, (ed. Smith S.) 180–181 (Wiley, 2012).
-
- Peterson R. & Nevalainen H. Trichoderma reesei RUT-C30–thirty years of strain improvement. Microbiology 158, 58–68 (2012). - PubMed
-
- Forment J. V., Ramón D. & MacCabe A. P. Consecutive gene deletions in Aspergillus nidulans: application of the Cre/loxP system. Curr. Genet. 50, 217–224 (2006). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

