CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation
- PMID: 26857612
- PMCID: PMC4746659
- DOI: 10.1038/srep20611
CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation
Abstract
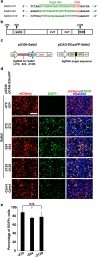
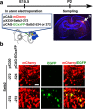
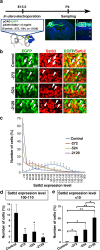
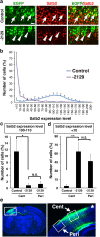
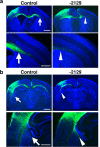
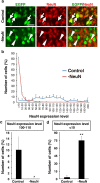
The CRISPR/Cas9 system has recently been adapted for generating knockout mice to investigate physiological functions and pathological mechanisms. Here, we report a highly efficient procedure for brain-specific disruption of genes of interest in vivo. We constructed pX330 plasmids expressing humanized Cas9 and single-guide RNAs (sgRNAs) against the Satb2 gene, which encodes an AT-rich DNA-binding transcription factor and is responsible for callosal axon projections in the developing mouse brain. We first confirmed that these constructs efficiently induced double-strand breaks (DSBs) in target sites of exogenous plasmids both in vitro and in vivo. We then found that the introduction of pX330-Satb2 into the developing mouse brain using in utero electroporation led to a dramatic reduction of Satb2 expression in the transfected cerebral cortex, suggesting DSBs had occurred in the Satb2 gene with high efficiency. Furthermore, we found that Cas9-mediated targeting of the Satb2 gene induced abnormalities in axonal projection patterns, which is consistent with the phenotypes previously observed in Satb2 mutant mice. Introduction of pX330-NeuN using our procedure also resulted in the efficient disruption of the NeuN gene. Thus, our procedure combining the CRISPR/Cas9 system and in utero electroporation is an effective and rapid approach to achieve brain-specific gene knockout in vivo.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

