Identification of HDAC Inhibitors Using a Cell-Based HDAC I/II Assay
- PMID: 26858181
- PMCID: PMC4917448
- DOI: 10.1177/1087057116629381
Identification of HDAC Inhibitors Using a Cell-Based HDAC I/II Assay
Abstract
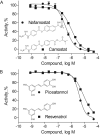
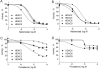
Histone deacetylases (HDACs) are a class of epigenetic enzymes that regulate gene expression by histone deacetylation. Altered HDAC function has been linked to cancer and neurodegenerative diseases, making HDACs popular therapeutic targets. In this study, we describe a screening approach for identification of compounds that inhibit endogenous class I and II HDACs. A homogeneous, luminogenic HDAC I/II assay was optimized in a 1536-well plate format in several human cancer cell lines, including HCT116 and human neural stem cells. The assay confirmed 37 known HDAC inhibitors from two libraries of known epigenetics-active compounds. Using the assay, we identified a group of potential HDAC inhibitors by screening the National Center for Advancing Translational Sciences (NCATS) Pharmaceutical Collection of 2527 small-molecule drugs. The selected compounds showed similar HDAC I/II inhibitory potency and efficacy values in both HCT116 and neural stem cells. Several previously unidentified HDAC inhibitors were further evaluated and profiled for their selectivity against a panel of 10 HDAC I/II isoforms using fluorogenic HDAC biochemical assays. In summary, our results show that several novel HDAC inhibitors, including nafamostat and piceatannol, have been identified using the HDAC I/II cell-based assay, and multiple cell types have been validated for high-throughput screening of large chemical libraries.
Keywords: cancer; epigenetics; histone deacetylase; neurodegenerative disease; qHTS.
© 2016 Society for Laboratory Automation and Screening.
Figures


References
-
- Berdasco M, Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum Genet. 2013;132(4):359–83. - PubMed
-
- Marmorstein R. Structure of histone deacetylases: insights into substrate recognition and catalysis. Structure. 2001;9(12):1127–33. - PubMed
-
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

