Metal-Dependent Function of a Mammalian Acireductone Dioxygenase
- PMID: 26858196
- PMCID: PMC5319410
- DOI: 10.1021/acs.biochem.5b01319
Metal-Dependent Function of a Mammalian Acireductone Dioxygenase
Abstract
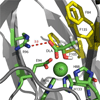
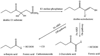
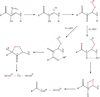
The two acireductone dioxygenase (ARD) isozymes from the methionine salvage pathway of Klebsiella oxytoca are the only known pair of naturally occurring metalloenzymes with distinct chemical and physical properties determined solely by the identity of the divalent transition metal ion (Fe(2+) or Ni(2+)) in the active site. We now show that this dual chemistry can also occur in mammals. ARD from Mus musculus (MmARD) was studied to relate the metal ion identity and three-dimensional structure to enzyme function. The iron-containing isozyme catalyzes the cleavage of 1,2-dihydroxy-3-keto-5-(thiomethyl)pent-1-ene (acireductone) by O2 to formate and the ketoacid precursor of methionine, which is the penultimate step in methionine salvage. The nickel-bound form of ARD catalyzes an off-pathway reaction resulting in formate, carbon monoxide (CO), and 3-(thiomethyl) propionate. Recombinant MmARD was expressed and purified to obtain a homogeneous enzyme with a single transition metal ion bound. The Fe(2+)-bound protein, which shows about 10-fold higher activity than that of others, catalyzes on-pathway chemistry, whereas the Ni(2+), Co(2+), or Mn(2+) forms exhibit off-pathway chemistry, as has been seen with ARD from Klebsiella. Thermal stability of the isozymes is strongly affected by the metal ion identity, with Ni(2+)-bound MmARD being the most stable, followed by Co(2+) and Fe(2+), and Mn(2+)-bound ARD being the least stable. Ni(2+)- and Co(2+)-bound MmARD were crystallized, and the structures of the two proteins found to be similar. Enzyme-ligand complexes provide insight into substrate binding, metal coordination, and the catalytic mechanism.
Figures


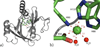




Similar articles
-
Dual chemistry catalyzed by human acireductone dioxygenase.Protein Eng Des Sel. 2017 Mar 1;30(3):197-204. doi: 10.1093/protein/gzw078. Protein Eng Des Sel. 2017. PMID: 28062648 Free PMC article.
-
A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).Biochemistry. 2020 Nov 10;59(44):4238-4249. doi: 10.1021/acs.biochem.0c00724. Epub 2020 Nov 2. Biochemistry. 2020. PMID: 33135413 Free PMC article.
-
The Metal Drives the Chemistry: Dual Functions of Acireductone Dioxygenase.Chem Rev. 2017 Aug 9;117(15):10474-10501. doi: 10.1021/acs.chemrev.7b00117. Epub 2017 Jul 21. Chem Rev. 2017. PMID: 28731690 Free PMC article. Review.
-
Characterization of metal binding in the active sites of acireductone dioxygenase isoforms from Klebsiella ATCC 8724.Biochemistry. 2008 Feb 26;47(8):2428-38. doi: 10.1021/bi7004152. Epub 2008 Feb 1. Biochemistry. 2008. PMID: 18237192 Free PMC article.
-
Ring-cleaving dioxygenases with a cupin fold.Appl Environ Microbiol. 2012 Apr;78(8):2505-14. doi: 10.1128/AEM.07651-11. Epub 2012 Jan 27. Appl Environ Microbiol. 2012. PMID: 22287012 Free PMC article. Review.
Cited by
-
Oxygen activation by mononuclear Mn, Co, and Ni centers in biology and synthetic complexes.J Biol Inorg Chem. 2017 Apr;22(2-3):407-424. doi: 10.1007/s00775-016-1402-7. Epub 2016 Nov 16. J Biol Inorg Chem. 2017. PMID: 27853875 Review.
-
Unraveling the Pivotal Roles of Various Metal Ion Centers in the Catalysis of Quercetin 2,4-Dioxygenases.Molecules. 2023 Aug 25;28(17):6238. doi: 10.3390/molecules28176238. Molecules. 2023. PMID: 37687067 Free PMC article.
-
Dual chemistry catalyzed by human acireductone dioxygenase.Protein Eng Des Sel. 2017 Mar 1;30(3):197-204. doi: 10.1093/protein/gzw078. Protein Eng Des Sel. 2017. PMID: 28062648 Free PMC article.
-
On the Structure and Reaction Mechanism of Human Acireductone Dioxygenase.Chemistry. 2018 Apr 6;24(20):5225-5237. doi: 10.1002/chem.201704617. Epub 2018 Jan 11. Chemistry. 2018. PMID: 29193386 Free PMC article.
-
A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).Biochemistry. 2020 Nov 10;59(44):4238-4249. doi: 10.1021/acs.biochem.0c00724. Epub 2020 Nov 2. Biochemistry. 2020. PMID: 33135413 Free PMC article.
References
-
- William-Ashman HG, Pegg AE. Polyamines in Biology and Medicine. New York: Marcel Dekker; 1981. pp. 43–73.
-
- Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5'-deoxy-5'-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. - PubMed
-
- Taiz L. Plant Physiology. Redwood City, CA: Benjamin/Cummins Publishin Co.; 1991.
-
- Schlenk F. Methylthioadenosine. Adv Enzymol Relat Areas Mol Biol. 1983;54:195–265. - PubMed
-
- Oredsson SM. Polyamine dependence of normal cell-cycle progression. Biochem Soc Trans. 2003;31:366–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

