Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis
- PMID: 26858247
- PMCID: PMC4817177
- DOI: 10.1074/jbc.M115.700344
Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis
Abstract
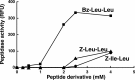
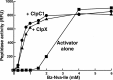
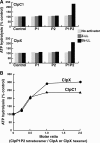
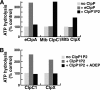
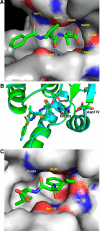
The ClpP protease complex and its regulatory ATPases, ClpC1 and ClpX, inMycobacterium tuberculosis(Mtb) are essential and, therefore, promising drug targets. TheMtbClpP protease consists of two heptameric rings, one composed of ClpP1 and the other of ClpP2 subunits. Formation of the enzymatically active ClpP1P2 complex requires binding of N-blocked dipeptide activators. We have found a new potent activator, benzoyl-leucine-leucine (Bz-LL), that binds with higher affinity and promotes 3-4-fold higher peptidase activity than previous activators. Bz-LL-activated ClpP1P2 specifically stimulates the ATPase activity ofMtbClpC1 and ClpX. The ClpC1P1P2 and ClpXP1P2 complexes exhibit 2-3-fold enhanced ATPase activity, peptide cleavage, and ATP-dependent protein degradation. The crystal structure of ClpP1P2 with bound Bz-LL was determined at a resolution of 3.07 Å and with benzyloxycarbonyl-Leu-Leu (Z-LL) bound at 2.9 Å. Bz-LL was present in all 14 active sites, whereas Z-LL density was not resolved. Surprisingly, Bz-LL adopts opposite orientations in ClpP1 and ClpP2. In ClpP1, Bz-LL binds with the C-terminal leucine side chain in the S1 pocket. One C-terminal oxygen is close to the catalytic serine, whereas the other contacts backbone amides in the oxyanion hole. In ClpP2, Bz-LL binds with the benzoyl group in the S1 pocket, and the peptide hydrogen bonded between parallel β-strands. The ClpP2 axial loops are extended, forming an open axial channel as has been observed with bound ADEP antibiotics. Thus occupancy of the active sites of ClpP allosterically alters sites on the surfaces thereby affecting the association of ClpP1 and ClpP2 rings, interactions with regulatory ATPases, and entry of protein substrates.
Keywords: enzyme mechanism; protease; protein complex; protein degradation; x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring.EMBO J. 2012 Mar 21;31(6):1529-41. doi: 10.1038/emboj.2012.5. Epub 2012 Jan 27. EMBO J. 2012. PMID: 22286948 Free PMC article.
-
The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1.PLoS One. 2015 May 1;10(5):e0125345. doi: 10.1371/journal.pone.0125345. eCollection 2015. PLoS One. 2015. PMID: 25933022 Free PMC article.
-
Cleavage Specificity of Mycobacterium tuberculosis ClpP1P2 Protease and Identification of Novel Peptide Substrates and Boronate Inhibitors with Anti-bacterial Activity.J Biol Chem. 2015 Apr 24;290(17):11008-20. doi: 10.1074/jbc.M114.625640. Epub 2015 Mar 10. J Biol Chem. 2015. PMID: 25759383 Free PMC article.
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
-
Regulatory proteins of the proteasome.Enzyme Protein. 1993;47(4-6):314-24. doi: 10.1159/000468689. Enzyme Protein. 1993. PMID: 7697129 Review.
Cited by
-
Acyldepsipeptide Analogues: A Future Generation Antibiotics for Tuberculosis Treatment.Pharmaceutics. 2022 Sep 15;14(9):1956. doi: 10.3390/pharmaceutics14091956. Pharmaceutics. 2022. PMID: 36145704 Free PMC article. Review.
-
Identification of the inhibitory mechanism of ecumicin and rufomycin 4-7 on the proteolytic activity of Mycobacterium tuberculosis ClpC1/ClpP1/ClpP2 complex.Tuberculosis (Edinb). 2023 Jan;138:102298. doi: 10.1016/j.tube.2022.102298. Epub 2022 Dec 24. Tuberculosis (Edinb). 2023. PMID: 36580851 Free PMC article.
-
Structural Insights into Bortezomib-Induced Activation of the Caseinolytic Chaperone-Protease System in Mycobacterium tuberculosis.Nat Commun. 2025 Apr 11;16(1):3466. doi: 10.1038/s41467-025-58410-4. Nat Commun. 2025. PMID: 40216758 Free PMC article.
-
Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.EMBO J. 2022 Jul 4;41(13):e109755. doi: 10.15252/embj.2021109755. Epub 2022 May 20. EMBO J. 2022. PMID: 35593068 Free PMC article.
-
Electrostatic interactions guide substrate recognition of the prokaryotic ubiquitin-like protein ligase PafA.Nat Commun. 2023 Aug 29;14(1):5266. doi: 10.1038/s41467-023-40807-8. Nat Commun. 2023. PMID: 37644028 Free PMC article.
References
-
- Porankiewicz J., Wang J., and Clarke A. K. (1999) New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32, 449–458 - PubMed
-
- Katayama-Fujimura Y., Gottesman S., and Maurizi M. R. (1987) A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262, 4477–4485 - PubMed
-
- Wang J., Hartling J. A., and Flanagan J. M. (1997) The structure of ClpP at 2.3 Å resolution suggests a model for ATP- dependent proteolysis. Cell 91, 447–456 - PubMed
-
- Kessel M., Maurizi M. R., Kim B., Kocsis E., Trus B. L., Singh S. K., and Steven A. C. (1995) Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J. Mol. Biol. 250, 587–594 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

