Balancing of B6 Vitamers Is Essential for Plant Development and Metabolism in Arabidopsis
- PMID: 26858304
- PMCID: PMC4790880
- DOI: 10.1105/tpc.15.01033
Balancing of B6 Vitamers Is Essential for Plant Development and Metabolism in Arabidopsis
Abstract
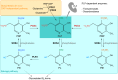
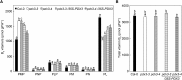
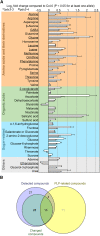
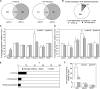
Vitamin B6 comprises a family of compounds that is essential for all organisms, most notable among which is the cofactor pyridoxal 5'-phosphate (PLP). Other forms of vitamin B6 include pyridoxamine 5'-phosphate (PMP), pyridoxine 5'-phosphate (PNP), and the corresponding nonphosphorylated derivatives. While plants can biosynthesize PLP de novo, they also have salvage pathways that serve to interconvert the different vitamers. The selective contribution of these various pathways to cellular vitamin B6 homeostasis in plants is not fully understood. Although biosynthesis de novo has been extensively characterized, the salvage pathways have received comparatively little attention in plants. Here, we show that the PMP/PNP oxidase PDX3 is essential for balancing B6 vitamer levels in Arabidopsis thaliana. In the absence of PDX3, growth and development are impaired and the metabolite profile is altered. Surprisingly, RNA sequencing reveals strong induction of stress-related genes in pdx3, particularly those associated with biotic stress that coincides with an increase in salicylic acid levels. Intriguingly, exogenous ammonium rescues the growth and developmental phenotype in line with a severe reduction in nitrate reductase activity that may be due to the overaccumulation of PMP in pdx3. Our analyses demonstrate an important link between vitamin B6 homeostasis and nitrogen metabolism.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures



Similar articles
-
Interaction between vitamin B6 metabolism, nitrogen metabolism and autoimmunity.Plant Signal Behav. 2016;11(4):e1161876. doi: 10.1080/15592324.2016.1161876. Plant Signal Behav. 2016. PMID: 27018849 Free PMC article.
-
Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5'-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway.Plant Physiol. 2007 Nov;145(3):985-96. doi: 10.1104/pp.107.105189. Epub 2007 Sep 14. Plant Physiol. 2007. PMID: 17873088 Free PMC article.
-
Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis.Plant Mol Biol. 2011 May;76(1-2):157-69. doi: 10.1007/s11103-011-9777-x. Epub 2011 May 1. Plant Mol Biol. 2011. PMID: 21533842
-
Pyridoxal 5'-Phosphate Biosynthesis by Pyridox-(am)-ine 5'-Phosphate Oxidase: Species-Specific Features.Int J Mol Sci. 2024 Mar 9;25(6):3174. doi: 10.3390/ijms25063174. Int J Mol Sci. 2024. PMID: 38542149 Free PMC article. Review.
-
Update on interconversions of vitamin B-6 with its coenzyme.J Nutr. 1999 Feb;129(2):325-7. doi: 10.1093/jn/129.2.325. J Nutr. 1999. PMID: 10024608 Review.
Cited by
-
Involvement of Pyridoxine/Pyridoxamine 5'-Phosphate Oxidase (PDX3) in Ethylene-Induced Auxin Biosynthesis in the Arabidopsis Root.Mol Cells. 2018 Dec 31;41(12):1033-1044. doi: 10.14348/molcells.2018.0363. Epub 2018 Nov 14. Mol Cells. 2018. PMID: 30453730 Free PMC article.
-
B vitamin supply in plants and humans: the importance of vitamer homeostasis.Plant J. 2022 Aug;111(3):662-682. doi: 10.1111/tpj.15859. Epub 2022 Jun 27. Plant J. 2022. PMID: 35673947 Free PMC article. Review.
-
Phosphorylated B6 vitamer deficiency in SALT OVERLY SENSITIVE 4 mutants compromises shoot and root development.Plant Physiol. 2022 Jan 20;188(1):220-240. doi: 10.1093/plphys/kiab475. Plant Physiol. 2022. PMID: 34730814 Free PMC article.
-
Metabolic Regulation and Lipidomic Remodeling in Relation to Spermidine-induced Stress Tolerance to High Temperature in Plants.Int J Mol Sci. 2022 Oct 13;23(20):12247. doi: 10.3390/ijms232012247. Int J Mol Sci. 2022. PMID: 36293104 Free PMC article.
-
Loss of a pyridoxal-phosphate phosphatase rescues Arabidopsis lacking an endoplasmic reticulum ATP carrier.Plant Physiol. 2022 May 3;189(1):49-65. doi: 10.1093/plphys/kiac048. Plant Physiol. 2022. PMID: 35139220 Free PMC article.
References
-
- Alcázar R., Parker J.E. (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16: 666–675. - PubMed
-
- Bilski P., Li M.Y., Ehrenshaft M., Daub M.E., Chignell C.F. (2000). Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 71: 129–134. - PubMed
-
- Bloom A.J. (2015). The increasing importance of distinguishing among plant nitrogen sources. Curr. Opin. Plant Biol. 25: 10–16. - PubMed
-
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

