Structure-function analysis of myomaker domains required for myoblast fusion
- PMID: 26858401
- PMCID: PMC4776501
- DOI: 10.1073/pnas.1600101113
Structure-function analysis of myomaker domains required for myoblast fusion
Abstract
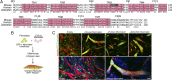
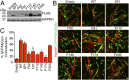
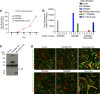
During skeletal muscle development, myoblasts fuse to form multinucleated myofibers. Myomaker [Transmembrane protein 8c (TMEM8c)] is a muscle-specific protein that is essential for myoblast fusion and sufficient to promote fusion of fibroblasts with muscle cells; however, the structure and biochemical properties of this membrane protein have not been explored. Here, we used CRISPR/Cas9 mutagenesis to disrupt myomaker expression in the C2C12 muscle cell line, which resulted in complete blockade to fusion. To define the functional domains of myomaker required to direct fusion, we established a heterologous cell-cell fusion system, in which fibroblasts expressing mutant versions of myomaker were mixed with WT myoblasts. Our data indicate that the majority of myomaker is embedded in the plasma membrane with seven membrane-spanning regions and a required intracellular C-terminal tail. We show that myomaker function is conserved in other mammalian orthologs; however, related family members (TMEM8a and TMEM8b) do not exhibit fusogenic activity. These findings represent an important step toward deciphering the cellular components and mechanisms that control myoblast fusion and muscle formation.
Keywords: CRISPR/Cas9; cell fusion; muscle development; myogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308(5720):369–373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL077439/HL/NHLBI NIH HHS/United States
- R01 DK099653/DK/NIDDK NIH HHS/United States
- DK-099653/DK/NIDDK NIH HHS/United States
- U01 HL100401/HL/NHLBI NIH HHS/United States
- R01 HL130253/HL/NHLBI NIH HHS/United States
- R01 HL111665/HL/NHLBI NIH HHS/United States
- HL-077439/HL/NHLBI NIH HHS/United States
- U01-HL-100401/HL/NHLBI NIH HHS/United States
- HL-111665/HL/NHLBI NIH HHS/United States
- HL-093039/HL/NHLBI NIH HHS/United States
- R01 AR067294/AR/NIAMS NIH HHS/United States
- R01 AR068286/AR/NIAMS NIH HHS/United States
- R01 HL093039/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

