Early and multiple origins of metastatic lineages within primary tumors
- PMID: 26858460
- PMCID: PMC4776530
- DOI: 10.1073/pnas.1525677113
Early and multiple origins of metastatic lineages within primary tumors
Abstract
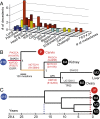
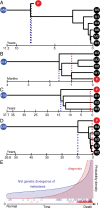
Many aspects of the evolutionary process of tumorigenesis that are fundamental to cancer biology and targeted treatment have been challenging to reveal, such as the divergence times and genetic clonality of metastatic lineages. To address these challenges, we performed tumor phylogenetics using molecular evolutionary models, reconstructed ancestral states of somatic mutations, and inferred cancer chronograms to yield three conclusions. First, in contrast to a linear model of cancer progression, metastases can originate from divergent lineages within primary tumors. Evolved genetic changes in cancer lineages likely affect only the proclivity toward metastasis. Single genetic changes are unlikely to be necessary or sufficient for metastasis. Second, metastatic lineages can arise early in tumor development, sometimes long before diagnosis. The early genetic divergence of some metastatic lineages directs attention toward research on driver genes that are mutated early in cancer evolution. Last, the temporal order of occurrence of driver mutations can be inferred from phylogenetic analysis of cancer chronograms, guiding development of targeted therapeutics effective against primary tumors and metastases.
Keywords: ancestral reconstruction; cancer; chronograms; oncogenes; tumor phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

