From chlorite dismutase towards HemQ - the role of the proximal H-bonding network in haeme binding
- PMID: 26858461
- PMCID: PMC4793301
- DOI: 10.1042/BSR20150330
From chlorite dismutase towards HemQ - the role of the proximal H-bonding network in haeme binding
Abstract
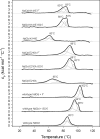
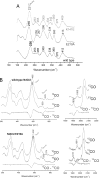
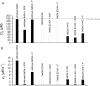
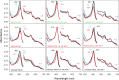
Chlorite dismutase (Cld) and HemQ are structurally and phylogenetically closely related haeme enzymes differing fundamentally in their enzymatic properties. Clds are able to convert chlorite into chloride and dioxygen, whereas HemQ is proposed to be involved in the haeme b synthesis of Gram-positive bacteria. A striking difference between these protein families concerns the proximal haeme cavity architecture. The pronounced H-bonding network in Cld, which includes the proximal ligand histidine and fully conserved glutamate and lysine residues, is missing in HemQ. In order to understand the functional consequences of this clearly evident difference, specific hydrogen bonds in Cld from 'Candidatus Nitrospira defluvii' (NdCld) were disrupted by mutagenesis. The resulting variants (E210A and K141E) were analysed by a broad set of spectroscopic (UV-vis, EPR and resonance Raman), calorimetric and kinetic methods. It is demonstrated that the haeme cavity architecture in these protein families is very susceptible to modification at the proximal site. The observed consequences of such structural variations include a significant decrease in thermal stability and also affinity between haeme b and the protein, a partial collapse of the distal cavity accompanied by an increased percentage of low-spin state for the E210A variant, lowered enzymatic activity concomitant with higher susceptibility to self-inactivation. The high-spin (HS) ligand fluoride is shown to exhibit a stabilizing effect and partially restore wild-type Cld structure and function. The data are discussed with respect to known structure-function relationships of Clds and the proposed function of HemQ as a coprohaeme decarboxylase in the last step of haeme biosynthesis in Firmicutes and Actinobacteria.
Keywords: H-bonding network; HemQ; chlorite dismutase; electron paramagnetic resonance spectroscopy; haeme binding; resonance Raman spectroscopy.
© 2016 Authors.
Figures





Similar articles
-
Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.Biochemistry. 2014 Jan 14;53(1):77-89. doi: 10.1021/bi401042z. Epub 2014 Jan 3. Biochemistry. 2014. PMID: 24364531 Free PMC article.
-
Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.FEBS J. 2016 Dec;283(23):4386-4401. doi: 10.1111/febs.13930. Epub 2016 Nov 14. FEBS J. 2016. PMID: 27758026 Free PMC article.
-
Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": identification of a catalytically important amino acid residue.J Struct Biol. 2010 Dec;172(3):331-42. doi: 10.1016/j.jsb.2010.06.014. Epub 2010 Jun 22. J Struct Biol. 2010. PMID: 20600954
-
HemQ: An iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria.Arch Biochem Biophys. 2015 May 15;574:27-35. doi: 10.1016/j.abb.2015.02.017. Epub 2015 Feb 21. Arch Biochem Biophys. 2015. PMID: 25711532 Free PMC article. Review.
-
Chlorite dismutases - a heme enzyme family for use in bioremediation and generation of molecular oxygen.Biotechnol J. 2014 Apr;9(4):461-73. doi: 10.1002/biot.201300210. Epub 2014 Feb 12. Biotechnol J. 2014. PMID: 24519858 Free PMC article. Review.
Cited by
-
A traffic light enzyme: acetate binding reversibly switches chlorite dismutase from a red- to a green-colored heme protein.J Biol Inorg Chem. 2020 Jun;25(4):609-620. doi: 10.1007/s00775-020-01784-1. Epub 2020 Apr 3. J Biol Inorg Chem. 2020. PMID: 32246282 Free PMC article.
-
Identification of a parasitic symbiosis between respiratory metabolisms in the biogeochemical chlorine cycle.ISME J. 2020 May;14(5):1194-1206. doi: 10.1038/s41396-020-0599-1. Epub 2020 Feb 5. ISME J. 2020. PMID: 32024948 Free PMC article.
-
Coproheme decarboxylases - Phylogenetic prediction versus biochemical experiments.Arch Biochem Biophys. 2018 Feb 15;640:27-36. doi: 10.1016/j.abb.2018.01.005. Epub 2018 Jan 10. Arch Biochem Biophys. 2018. PMID: 29331688 Free PMC article.
-
Reactivity of Coproheme Decarboxylase with Monovinyl, Monopropionate Deuteroheme.Biomolecules. 2023 Jun 6;13(6):946. doi: 10.3390/biom13060946. Biomolecules. 2023. PMID: 37371526 Free PMC article.
-
Reaction intermediate rotation during the decarboxylation of coproheme to heme b in C. diphtheriae.Biophys J. 2021 Sep 7;120(17):3600-3614. doi: 10.1016/j.bpj.2021.06.042. Epub 2021 Jul 31. Biophys J. 2021. PMID: 34339636 Free PMC article.
References
-
- Hofbauer S., Hagmüller A., Schaffner I., Mlynek G., Krutzler M., Stadlmayr G., Pirker K.F., Obinger C., Daims H., Djinović-Carugo K., Furtmüller P.G. Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes. Arch. Biochem. Biophys. 2015;574:36–48. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

