High-Throughput Isolation of Giant Viruses in Liquid Medium Using Automated Flow Cytometry and Fluorescence Staining
- PMID: 26858703
- PMCID: PMC4731542
- DOI: 10.3389/fmicb.2016.00026
High-Throughput Isolation of Giant Viruses in Liquid Medium Using Automated Flow Cytometry and Fluorescence Staining
Abstract
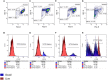
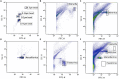
The isolation of giant viruses using amoeba co-culture is tedious and fastidious. Recently, the procedure was successfully associated with a method that detects amoebal lysis on agar plates. However, the procedure remains time-consuming and is limited to protozoa growing on agar. We present here advances for the isolation of giant viruses. A high-throughput automated method based on flow cytometry and fluorescent staining was used to detect the presence of giant viruses in liquid medium. Development was carried out with the Acanthamoeba polyphaga strain widely used in past and current co-culture experiments. The proof of concept was validated with virus suspensions: artificially contaminated samples but also environmental samples from which viruses were previously isolated. After validating the technique, and fortuitously isolating a new Mimivirus, we automated the technique on 96-well plates and tested it on clinical and environmental samples using other protozoa. This allowed us to detect more than 10 strains of previously known species of giant viruses and seven new strains of a new virus lineage. This automated high-throughput method demonstrated significant time saving, and higher sensitivity than older techniques. It thus creates the means to isolate giant viruses at high speed.
Keywords: automated system; flow cytometry; fluorescence staining; gating strategy; giant viruses; high-throughput; protozoa.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

