The Global Reciprocal Reprogramming between Mycobacteriophage SWU1 and Mycobacterium Reveals the Molecular Strategy of Subversion and Promotion of Phage Infection
- PMID: 26858712
- PMCID: PMC4729954
- DOI: 10.3389/fmicb.2016.00041
The Global Reciprocal Reprogramming between Mycobacteriophage SWU1 and Mycobacterium Reveals the Molecular Strategy of Subversion and Promotion of Phage Infection
Abstract
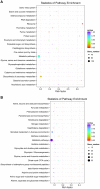
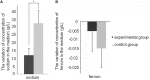
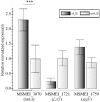
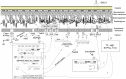
Bacteriophages are the viruses of bacteria, which have contributed extensively to our understanding of life and modern biology. The phage-mediated bacterial growth inhibition represents immense untapped source for novel antimicrobials. Insights into the interaction between mycobacteriophage and Mycobacterium host will inform better utilizing of mycobacteriophage. In this study, RNA sequencing technology (RNA-seq) was used to explore the global response of Mycobacterium smegmatis mc(2)155 at an early phase of infection with mycobacteriophage SWU1, key host metabolic processes of M. smegmatis mc(2)155 shut off by SWU1, and the responsible phage proteins. The results of RNA-seq were confirmed by Real-time PCR and functional assay. 1174 genes of M. smegmatis mc(2)155 (16.9% of the entire encoding capacity) were differentially regulated by phage infection. These genes belong to six functional categories: (i) signal transduction, (ii) cell energetics, (iii) cell wall biosynthesis, (iv) DNA, RNA, and protein biosynthesis, (v) iron uptake, (vi) central metabolism. The transcription patterns of phage SWU1 were also characterized. This study provided the first global glimpse of the reciprocal reprogramming between the mycobacteriophage and Mycobacterium host.
Keywords: interaction; ion channel; mycobacteriophage; mycobacterium; transcriptome.
Figures

Similar articles
-
Insertion Mutation of MSMEG_0392 Play an Important Role in Resistance of M. smegmatis to Mycobacteriophage SWU1.Infect Drug Resist. 2022 Feb 2;15:347-357. doi: 10.2147/IDR.S341494. eCollection 2022. Infect Drug Resist. 2022. PMID: 35140480 Free PMC article.
-
Genomic and proteomic features of mycobacteriophage SWU1 isolated from China soil.Gene. 2015 Apr 25;561(1):45-53. doi: 10.1016/j.gene.2015.02.053. Epub 2015 Feb 19. Gene. 2015. PMID: 25701596 Free PMC article.
-
Mycobacteriophage SWU1-Functionalized magnetic particles for facile bioluminescent detection of Mycobacterium smegmatis.Anal Chim Acta. 2021 Feb 8;1145:17-25. doi: 10.1016/j.aca.2020.12.009. Epub 2020 Dec 8. Anal Chim Acta. 2021. PMID: 33453875
-
Biology of a novel mycobacteriophage, SWU1, isolated from Chinese soil as revealed by genomic characteristics.J Virol. 2012 Sep;86(18):10230-1. doi: 10.1128/JVI.01568-12. J Virol. 2012. PMID: 22923793 Free PMC article.
-
Mycobacteriophage SWU1 gp39 can potentiate multiple antibiotics against Mycobacterium via altering the cell wall permeability.Sci Rep. 2016 Jun 28;6:28701. doi: 10.1038/srep28701. Sci Rep. 2016. PMID: 27350398 Free PMC article.
Cited by
-
Mycobacteriophages.Microbiol Spectr. 2018 Oct;6(5):10.1128/microbiolspec.gpp3-0026-2018. doi: 10.1128/microbiolspec.GPP3-0026-2018. Microbiol Spectr. 2018. PMID: 30291704 Free PMC article. Review.
-
Transcriptional dynamics during Rhodococcus erythropolis infection with phage WC1.BMC Microbiol. 2024 Apr 1;24(1):107. doi: 10.1186/s12866-024-03241-4. BMC Microbiol. 2024. PMID: 38561651 Free PMC article.
-
Expression and evolutionary patterns of mycobacteriophage D29 and its temperate close relatives.BMC Microbiol. 2017 Dec 2;17(1):225. doi: 10.1186/s12866-017-1131-2. BMC Microbiol. 2017. PMID: 29197343 Free PMC article.
-
Isolation and Characterization of the Novel Phage JD032 and Global Transcriptomic Response during JD032 Infection of Clostridioides difficile Ribotype 078.mSystems. 2020 May 5;5(3):e00017-20. doi: 10.1128/mSystems.00017-20. mSystems. 2020. PMID: 32371470 Free PMC article.
-
A Novel Inovirus Reprograms Metabolism and Motility of Marine Alteromonas.Microbiol Spectr. 2022 Dec 21;10(6):e0338822. doi: 10.1128/spectrum.03388-22. Epub 2022 Oct 27. Microbiol Spectr. 2022. PMID: 36301121 Free PMC article.
References
-
- Belanger A. E., Besra G. S., Ford M. E., Mikusová K., Belisle J. T., Brennan P. J., et al. (1996). The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. U.S.A. 93, 11919–11924. 10.1073/pnas.93.21.11919 - DOI - PMC - PubMed
-
- Boulanger P., Letellier L. (1992). Ion channels are likely to be involved in the two steps of phage T5 DNA penetration into Escherichia coli cells. J. Biol. Chem. 267, 3168–3172. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

