A method for analyzing multiple continuous phenotypes in rare variant association studies allowing for flexible correlations in variant effects
- PMID: 26860061
- PMCID: PMC4989219
- DOI: 10.1038/ejhg.2016.8
A method for analyzing multiple continuous phenotypes in rare variant association studies allowing for flexible correlations in variant effects
Abstract
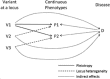
For region-based sequencing data, power to detect genetic associations can be improved through analysis of multiple related phenotypes. With this motivation, we propose a novel test to detect association simultaneously between a set of rare variants, such as those obtained by sequencing in a small genomic region, and multiple continuous phenotypes. We allow arbitrary correlations among the phenotypes and build on a linear mixed model by assuming the effects of the variants follow a multivariate normal distribution with a zero mean and a specific covariance matrix structure. In order to account for the unknown correlation parameter in the covariance matrix of the variant effects, a data-adaptive variance component test based on score-type statistics is derived. As our approach can calculate the P-value analytically, the proposed test procedure is computationally efficient. Broad simulations and an application to the UK10K project show that our proposed multivariate test is generally more powerful than univariate tests, especially when there are pleiotropic effects or highly correlated phenotypes.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

