Quorum sensing activity of Citrobacter amalonaticus L8A, a bacterium isolated from dental plaque
- PMID: 26860259
- PMCID: PMC4748228
- DOI: 10.1038/srep20702
Quorum sensing activity of Citrobacter amalonaticus L8A, a bacterium isolated from dental plaque
Abstract
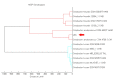
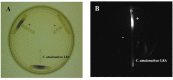
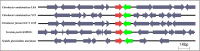
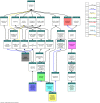
Cell-cell communication is also known as quorum sensing (QS) that happens in the bacterial cells with the aim to regulate their genes expression in response to increased cell density. In this study, a bacterium (L8A) isolated from dental plaque biofilm was identified as Citrobacter amalonaticus by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS). Its N-acylhomoserine-lactone (AHL) production was screened by using two types of AHL biosensors namely Chromobacterium violaceum CV026 and Escherichia coli [pSB401]. Citrobacter amalonaticus strain L8A was identified and confirmed producing numerous types of AHL namely N-butyryl-L-homoserine lactone (C4-HSL), N-hexanoyl-L-homoserine lactone (C6-HSL), N-octanoyl-L-homoserine lactone (C8-HSL) and N-hexadecanoyl-L-homoserine lactone (C16-HSL). We performed the whole genome sequence analysis of this oral isolate where its genome sequence reveals the presence of QS signal synthase gene and our work will pave the ways to study the function of the related QS genes in this bacterium.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Characterization of quorum sensing genes and N-acyl homoserine lactones in Citrobacter amalonaticus strain YG6.Gene. 2019 Feb 5;684:58-69. doi: 10.1016/j.gene.2018.10.031. Epub 2018 Oct 12. Gene. 2019. PMID: 30321658
-
Production of N-acyl homoserine lactones by Chromobacterium haemolyticum KM2 isolated from the river water in Malaysia.Arch Microbiol. 2018 Sep;200(7):1135-1142. doi: 10.1007/s00203-018-1526-y. Epub 2018 May 23. Arch Microbiol. 2018. PMID: 29796703
-
Acyl-Homoserine Lactone Production in Nitrifying Bacteria of the Genera Nitrosospira, Nitrobacter, and Nitrospira Identified via a Survey of Putative Quorum-Sensing Genes.Appl Environ Microbiol. 2017 Oct 31;83(22):e01540-17. doi: 10.1128/AEM.01540-17. Print 2017 Nov 15. Appl Environ Microbiol. 2017. PMID: 28887424 Free PMC article.
-
Quorum sensing and expression of virulence in pectobacteria.Sensors (Basel). 2012;12(3):3327-49. doi: 10.3390/s120303327. Epub 2012 Mar 8. Sensors (Basel). 2012. PMID: 22737011 Free PMC article. Review.
-
Quorum sensing: fact, fiction, and everything in between.Adv Appl Microbiol. 2007;62:191-234. doi: 10.1016/S0065-2164(07)62007-3. Adv Appl Microbiol. 2007. PMID: 17869606 Free PMC article. Review. No abstract available.
Cited by
-
Quorum-Sensing Inhibition by Gram-Positive Bacteria.Microorganisms. 2022 Feb 3;10(2):350. doi: 10.3390/microorganisms10020350. Microorganisms. 2022. PMID: 35208805 Free PMC article. Review.
-
Acyl homoserine lactone-mediated quorum sensing in the oral cavity: a paradigm revisited.Sci Rep. 2020 Jun 17;10(1):9800. doi: 10.1038/s41598-020-66704-4. Sci Rep. 2020. PMID: 32555242 Free PMC article.
-
The quorum quenching enzyme Aii20J modifies in vitro periodontal biofilm formation.Front Cell Infect Microbiol. 2023 Feb 2;13:1118630. doi: 10.3389/fcimb.2023.1118630. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36816581 Free PMC article.
-
Formicincola oecophyllae gen. nov. sp. nov., a novel member of the family Acetobacteraceae isolated from the weaver ant Oecophylla smaragdina.Antonie Van Leeuwenhoek. 2022 Aug;115(8):995-1007. doi: 10.1007/s10482-022-01750-8. Epub 2022 Jun 8. Antonie Van Leeuwenhoek. 2022. PMID: 35674967
-
Silent signals: how N-acyl homoserine lactones drive oral microbial behaviour and health outcomes.Front Oral Health. 2024 Dec 5;5:1484005. doi: 10.3389/froh.2024.1484005. eCollection 2024. Front Oral Health. 2024. PMID: 39703871 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

