Identification of Conserved and Diverse Metabolic Shifts during Rice Grain Development
- PMID: 26860358
- PMCID: PMC4748235
- DOI: 10.1038/srep20942
Identification of Conserved and Diverse Metabolic Shifts during Rice Grain Development
Abstract
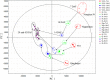
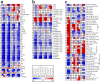
Seed development dedicates to reserve synthesis and accumulation and uncovering its genetic and biochemical mechanisms has been a major research focus. Although proteomic and transcriptomic analyses revealed dynamic changes of genes and enzymes involved, the information regarding concomitant metabolic changes is missing. Here we investigated the dynamic metabolic changes along the rice grain development of two japonica and two indica cultivars using non-targeted metabolomics approach, in which we successfully identified 214 metabolites. Statistical analyses revealed both cultivar and developmental stage dependent metabolic changes in rice grains. Generally, the stage specific metabolic kinetics corresponded well to the physiological status of the developing grains, and metabolic changes in developing rice grain are similar to those of dicot Arabidopsis and tomato at reserve accumulation stage but are different from those of dicots at seed desiccation stage. The remarkable difference in metabolite abundances between japonica and indica rice grain was observed at the reserve accumulation stage. Metabolite-metabolite correlation analysis uncovered potential new pathways for several metabolites. Taken together, this study uncovered both conserved and diverse development associated metabolic kinetics of rice grains, which facilitates further study to explore fundamental questions regarding the evolution of seed metabolic capabilities as well as their potential applications in crop improvement.
Figures




References
-
- Itoh J. I. et al. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46, 23–47 (2005). - PubMed
-
- Liu T., Tausta S. L., Gandotra N. & Nelson T. Rice Seed Development: Highly Resolved Transcriptomic Views. In: Agrawal G. K. & Rakwal R. Eds. Seed Development: OMICS Technologies toward Improvement of Seed Quality and Crop Yield, 61–80 (Springer, New York, 2012).
-
- Huh S. M. et al. Comparative transcriptome profiling of developing caryopses from two rice cultivars with differential dormancy. J. Plant Physiol. 170, 1090–1100 (2013). - PubMed
-
- Xu H. et al. Proteomic analysis of embryo development in rice (Oryza sativa). Planta 235, 687–701 (2012). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

