Mapping the Interaction Network of Key Proteins Involved in Histone mRNA Generation: A Hydrogen/Deuterium Exchange Study
- PMID: 26860583
- PMCID: PMC4847949
- DOI: 10.1016/j.jmb.2016.01.031
Mapping the Interaction Network of Key Proteins Involved in Histone mRNA Generation: A Hydrogen/Deuterium Exchange Study
Abstract
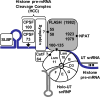
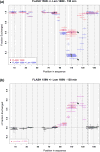
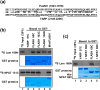
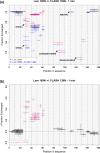
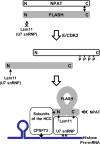
Histone pre-mRNAs are cleaved at the 3' end by a complex that contains U7 snRNP, the FLICE-associated huge protein (FLASH) and histone pre-mRNA cleavage complex (HCC) consisting of several polyadenylation factors. Within the complex, the N terminus of FLASH interacts with the N terminus of the U7 snRNP protein Lsm11, and together they recruit the HCC. FLASH through its distant C terminus independently interacts with the C-terminal SANT/Myb-like domain of nuclear protein, ataxia-telangiectasia locus (NPAT), a transcriptional co-activator required for expression of histone genes in S phase. To gain structural information on these interactions, we used mass spectrometry to monitor hydrogen/deuterium exchange in various regions of FLASH, Lsm11 and NPAT alone or in the presence of their respective binding partners. Our results indicate that the FLASH-interacting domain in Lsm11 is highly dynamic, while the more downstream region required for recruiting the HCC exchanges deuterium slowly and likely folds into a stable structure. In FLASH, a stable structure is adopted by the domain that interacts with Lsm11 and this domain is further stabilized by binding Lsm11. Notably, both hydrogen/deuterium exchange experiments and in vitro binding assays demonstrate that Lsm11, in addition to interacting with the N-terminal region of FLASH, also contacts the C-terminal SANT/Myb-like domain of FLASH, the same region that binds NPAT. However, while NPAT stabilizes this domain, Lsm11 causes its partial relaxation. These competing reactions may play a role in regulating histone gene expression in vivo.
Keywords: FLASH; Lsm11; Mass spectrometry; NPAT; U7 snRNP.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein.RNA. 2017 Jun;23(6):938-951. doi: 10.1261/rna.060806.117. Epub 2017 Mar 13. RNA. 2017. PMID: 28289156 Free PMC article.
-
3'-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors.RNA. 2013 Dec;19(12):1726-44. doi: 10.1261/rna.040360.113. Epub 2013 Oct 21. RNA. 2013. PMID: 24145821 Free PMC article.
-
Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila.RNA. 2011 Jun;17(6):1132-47. doi: 10.1261/rna.2566811. Epub 2011 Apr 27. RNA. 2011. PMID: 21525146 Free PMC article.
-
Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body.RNA Biol. 2017 Jun 3;14(6):726-738. doi: 10.1080/15476286.2016.1265198. Epub 2017 Jan 6. RNA Biol. 2017. PMID: 28059623 Free PMC article. Review.
-
Birth and Death of Histone mRNAs.Trends Genet. 2017 Oct;33(10):745-759. doi: 10.1016/j.tig.2017.07.014. Epub 2017 Aug 31. Trends Genet. 2017. PMID: 28867047 Free PMC article. Review.
Cited by
-
A region of SLBP outside the mRNA-processing domain is essential for deposition of histone mRNA into the Drosophila egg.J Cell Sci. 2021 Feb 11;134(3):jcs251728. doi: 10.1242/jcs.251728. J Cell Sci. 2021. PMID: 33408246 Free PMC article.
-
SUMO2 promotes histone pre-mRNA processing by stabilizing histone locus body interactions and facilitating U7 snRNP assembly.Genes Dev. 2025 Jul 10:10.1101/gad.352728.125. doi: 10.1101/gad.352728.125. Online ahead of print. Genes Dev. 2025. PMID: 40639911 Free PMC article.
-
Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations.Int J Mol Sci. 2020 Jul 24;21(15):5268. doi: 10.3390/ijms21155268. Int J Mol Sci. 2020. PMID: 32722282 Free PMC article.
-
U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein.RNA. 2017 Jun;23(6):938-951. doi: 10.1261/rna.060806.117. Epub 2017 Mar 13. RNA. 2017. PMID: 28289156 Free PMC article.
-
The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3'-end processing.PLoS One. 2017 Oct 11;12(10):e0186034. doi: 10.1371/journal.pone.0186034. eCollection 2017. PLoS One. 2017. PMID: 29020104 Free PMC article.
References
-
- Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. - PubMed
-
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, Van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

