Co-immunoprecipitation with Tau Isoform-specific Antibodies Reveals Distinct Protein Interactions and Highlights a Putative Role for 2N Tau in Disease
- PMID: 26861879
- PMCID: PMC4825019
- DOI: 10.1074/jbc.M115.641902
Co-immunoprecipitation with Tau Isoform-specific Antibodies Reveals Distinct Protein Interactions and Highlights a Putative Role for 2N Tau in Disease
Abstract
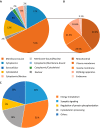
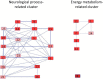
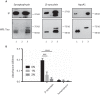
Alternative splicing generates multiple isoforms of the microtubule-associated protein Tau, but little is known about their specific function. In the adult mouse brain, three Tau isoforms are expressed that contain either 0, 1, or 2 N-terminal inserts (0N, 1N, and 2N). We generated Tau isoform-specific antibodies and performed co-immunoprecipitations followed by tandem mass tag multiplexed quantitative mass spectrometry. We identified novel Tau-interacting proteins of which one-half comprised membrane-bound proteins, localized to the plasma membrane, mitochondria, and other organelles. Tau was also found to interact with proteins involved in presynaptic signal transduction. MetaCore analysis revealed one major Tau interaction cluster that contained 33 Tau pulldown proteins. To explore the pathways in which these proteins are involved, we conducted an ingenuity pathway analysis that revealed two significant overlapping pathways, "cell-to-cell signaling and interaction" and "neurological disease." The functional enrichment tool DAVID showed that in particular the 2N Tau-interacting proteins were specifically associated with neurological disease. Finally, for a subset of Tau interactions (apolipoprotein A1 (apoA1), apoE, mitochondrial creatine kinase U-type, β-synuclein, synaptogyrin-3, synaptophysin, syntaxin 1B, synaptotagmin, and synapsin 1), we performed reverse co-immunoprecipitations, confirming the preferential interaction of specific isoforms. For example, apoA1 displayed a 5-fold preference for the interaction with 2N, whereas β-synuclein showed preference for 0N. Remarkably, a reverse immunoprecipitation with apoA1 detected only the 2N isoform. This highlights distinct protein interactions of the different Tau isoforms, suggesting that they execute different functions in brain tissue.
Keywords: Alzheimer disease; Tau protein (Tau); Tauopathy; apolipoprotein; protein/protein interaction.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Matus A. (1990) Microtubule-associated proteins and the determination of neuronal form. J. Physiol. 84, 134–137 - PubMed
-
- Tashiro K., Hasegawa M., Ihara Y., and Iwatsubo T. (1997) Somatodendritic localization of phosphorylated Tau in neonatal and adult rat cerebral cortex. Neuroreport 8, 2797–2801 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

