Oligonucleotide-Mediated Genome Editing Provides Precision and Function to Engineered Nucleases and Antibiotics in Plants
- PMID: 26864017
- PMCID: PMC4825113
- DOI: 10.1104/pp.15.01696
Oligonucleotide-Mediated Genome Editing Provides Precision and Function to Engineered Nucleases and Antibiotics in Plants
Abstract
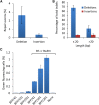
Here, we report a form of oligonucleotide-directed mutagenesis for precision genome editing in plants that uses single-stranded oligonucleotides (ssODNs) to precisely and efficiently generate genome edits at DNA strand lesions made by DNA double strand break reagents. Employing a transgene model in Arabidopsis (Arabidopsis thaliana), we obtained a high frequency of precise targeted genome edits when ssODNs were introduced into protoplasts that were pretreated with the glycopeptide antibiotic phleomycin, a nonspecific DNA double strand breaker. Simultaneous delivery of ssODN and a site-specific DNA double strand breaker, either transcription activator-like effector nucleases (TALENs) or clustered, regularly interspaced, short palindromic repeats (CRISPR/Cas9), resulted in a much greater targeted genome-editing frequency compared with treatment with DNA double strand-breaking reagents alone. Using this site-specific approach, we applied the combination of ssODN and CRISPR/Cas9 to develop an herbicide tolerance trait in flax (Linum usitatissimum) by precisely editing the 5'-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) genes. EPSPS edits occurred at sufficient frequency that we could regenerate whole plants from edited protoplasts without employing selection. These plants were subsequently determined to be tolerant to the herbicide glyphosate in greenhouse spray tests. Progeny (C1) of these plants showed the expected Mendelian segregation of EPSPS edits. Our findings show the enormous potential of using a genome-editing platform for precise, reliable trait development in crop plants.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

