Regulatory T Cells in Melanoma Revisited by a Computational Clustering of FOXP3+ T Cell Subpopulations
- PMID: 26864030
- PMCID: PMC4777917
- DOI: 10.4049/jimmunol.1402695
Regulatory T Cells in Melanoma Revisited by a Computational Clustering of FOXP3+ T Cell Subpopulations
Abstract
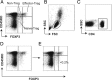
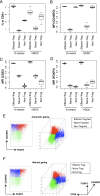
CD4(+) T cells that express the transcription factor FOXP3 (FOXP3(+) T cells) are commonly regarded as immunosuppressive regulatory T cells (Tregs). FOXP3(+) T cells are reported to be increased in tumor-bearing patients or animals and are considered to suppress antitumor immunity, but the evidence is often contradictory. In addition, accumulating evidence indicates that FOXP3 is induced by antigenic stimulation and that some non-Treg FOXP3(+) T cells, especially memory-phenotype FOXP3(low) cells, produce proinflammatory cytokines. Accordingly, the subclassification of FOXP3(+) T cells is fundamental for revealing the significance of FOXP3(+) T cells in tumor immunity, but the arbitrariness and complexity of manual gating have complicated the issue. In this article, we report a computational method to automatically identify and classify FOXP3(+) T cells into subsets using clustering algorithms. By analyzing flow cytometric data of melanoma patients, the proposed method showed that the FOXP3(+) subpopulation that had relatively high FOXP3, CD45RO, and CD25 expressions was increased in melanoma patients, whereas manual gating did not produce significant results on the FOXP3(+) subpopulations. Interestingly, the computationally identified FOXP3(+) subpopulation included not only classical FOXP3(high) Tregs, but also memory-phenotype FOXP3(low) cells by manual gating. Furthermore, the proposed method successfully analyzed an independent data set, showing that the same FOXP3(+) subpopulation was increased in melanoma patients, validating the method. Collectively, the proposed method successfully captured an important feature of melanoma without relying on the existing criteria of FOXP3(+) T cells, revealing a hidden association between the T cell profile and melanoma, and providing new insights into FOXP3(+) T cells and Tregs.
Copyright © 2016 The Authors.
Figures




References
-
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. 2008. Regulatory T cells and immune tolerance. Cell 133: 775–787. - PubMed
-
- Zou W. 2006. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 6: 295–307. - PubMed
-
- deLeeuw, R. J., S. E. Kost, J. A. Kakal, and B. H. Nelson. 2012. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin. Cancer Res. 18: 3022–3029. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

