Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM
- PMID: 26864203
- PMCID: PMC4780610
- DOI: 10.1073/pnas.1517883113
Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM
Abstract
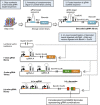
The orchestrated action of genes controls complex biological phenotypes, yet the systematic discovery of gene and drug combinations that modulate these phenotypes in human cells is labor intensive and challenging to scale. Here, we created a platform for the massively parallel screening of barcoded combinatorial gene perturbations in human cells and translated these hits into effective drug combinations. This technology leverages the simplicity of the CRISPR-Cas9 system for multiplexed targeting of specific genomic loci and the versatility of combinatorial genetics en masse (CombiGEM) to rapidly assemble barcoded combinatorial genetic libraries that can be tracked with high-throughput sequencing. We applied CombiGEM-CRISPR to create a library of 23,409 barcoded dual guide-RNA (gRNA) combinations and then perform a high-throughput pooled screen to identify gene pairs that inhibited ovarian cancer cell growth when they were targeted. We validated the growth-inhibiting effects of specific gene sets, including epigenetic regulators KDM4C/BRD4 and KDM6B/BRD4, via individual assays with CRISPR-Cas-based knockouts and RNA-interference-based knockdowns. We also tested small-molecule drug pairs directed against our pairwise hits and showed that they exerted synergistic antiproliferative effects against ovarian cancer cells. We envision that the CombiGEM-CRISPR platform will be applicable to a broad range of biological settings and will accelerate the systematic identification of genetic combinations and their translation into novel drug combinations that modulate complex human disease phenotypes.
Keywords: CRISPR-Cas; CombiGEM; genetic perturbations; high-throughput screening; multifactorial genetics.
Conflict of interest statement
Conflict of interest statement: T.K.L., A.S.L.W., and G.C.G.C. have filed a patent application based on this work with the US Patent and Trademark Office.
Figures


References
-
- Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30(7):679–692. - PubMed
-
- Kummar S, et al. Utilizing targeted cancer therapeutic agents in combination: Novel approaches and urgent requirements. Nat Rev Drug Discov. 2010;9(11):843–856. - PubMed
-
- Holbeck S. 2014 Cell-based screens of drug combinations at NCI. National Institutes of Health. Available at deainfo.nci.nih.gov/advisory/ctac/1114/5%20-%20Holbeck.pdf. Accessed January 14, 2016.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

