De novo construction of a "Gene-space" for diploid plant genome rich in repetitive sequences by an iterative Process of Extraction and Assembly of NGS reads (iPEA protocol) with limited computing resources
- PMID: 26864345
- PMCID: PMC4750290
- DOI: 10.1186/s13104-016-1903-z
De novo construction of a "Gene-space" for diploid plant genome rich in repetitive sequences by an iterative Process of Extraction and Assembly of NGS reads (iPEA protocol) with limited computing resources
Abstract
Background: The continuing increase in size and quality of the "short reads" raw data is a significant help for the quality of the assembly obtained through various bioinformatics tools. However, building a reference genome sequence for most plant species remains a significant challenge due to the large number of repeated sequences which are problematic for a whole-genome quality de novo assembly. Furthermore, for most SNP identification approaches in plant genetics and breeding, only the "Gene-space" regions including the promoter, exon and intron sequences are considered.
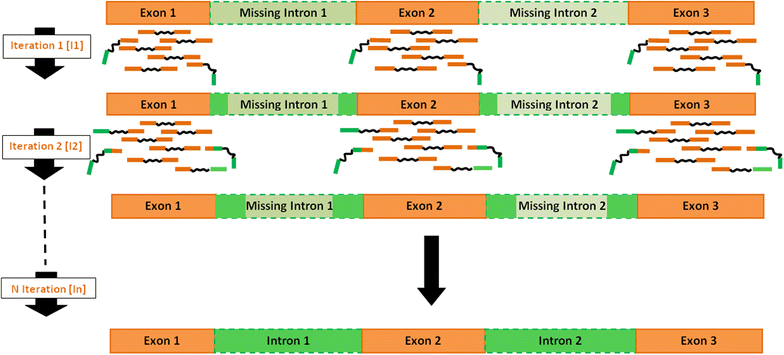
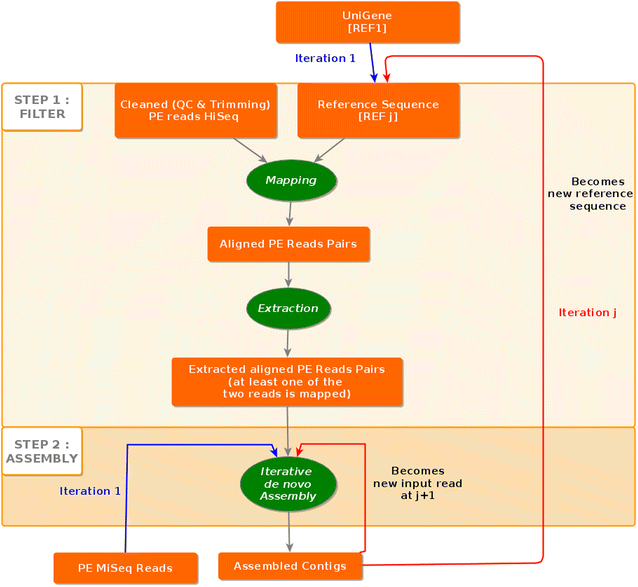
Results: We developed the iPea protocol to produce a de novo Gene-space assembly by reconstructing, in an iterative way, the non-coding sequence flanking the Unigene cDNA sequence through addition of next-generation DNA-seq data. The approach was elaborated with the large diploid genome of pea (Pisum sativum L.), rich in repetitive sequences. The final Gene-space assembly included 35,400 contigs (97 Mb), covering 88 % of the 40,227 contigs (53.1 Mb) of the PsCam_low-copy Unigen set. Its accuracy was validated by the results of the built GenoPea 13.2 K SNP Array.
Conclusion: The iPEA protocol allows the reconstruction of a Gene-space based from RNA-Seq and DNA-seq data with limited computing resources.
Figures



Similar articles
-
Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species.Plant J. 2015 Oct;84(1):1-19. doi: 10.1111/tpj.12967. Plant J. 2015. PMID: 26296678
-
Development of two major resources for pea genomics: the GenoPea 13.2K SNP Array and a high-density, high-resolution consensus genetic map.Plant J. 2015 Dec;84(6):1257-73. doi: 10.1111/tpj.13070. Plant J. 2015. PMID: 26590015
-
RepAHR: an improved approach for de novo repeat identification by assembly of the high-frequency reads.BMC Bioinformatics. 2020 Oct 19;21(1):463. doi: 10.1186/s12859-020-03779-w. BMC Bioinformatics. 2020. PMID: 33076827 Free PMC article.
-
The present and future of de novo whole-genome assembly.Brief Bioinform. 2018 Jan 1;19(1):23-40. doi: 10.1093/bib/bbw096. Brief Bioinform. 2018. PMID: 27742661 Review.
-
Effects of genome structure variation, homeologous genes and repetitive DNA on polyploid crop research in the age of genomics.Plant Sci. 2016 Jan;242:37-46. doi: 10.1016/j.plantsci.2015.09.017. Epub 2015 Sep 26. Plant Sci. 2016. PMID: 26566823 Review.
Cited by
-
Quick and efficient approach to develop genomic resources in orphan species: Application in Lavandula angustifolia.PLoS One. 2020 Dec 11;15(12):e0243853. doi: 10.1371/journal.pone.0243853. eCollection 2020. PLoS One. 2020. PMID: 33306734 Free PMC article.
-
Virtual Genome Walking across the 32 Gb Ambystoma mexicanum genome; assembling gene models and intronic sequence.Sci Rep. 2018 Jan 12;8(1):618. doi: 10.1038/s41598-017-19128-6. Sci Rep. 2018. PMID: 29330416 Free PMC article.
-
HopBase: a unified resource for Humulus genomics.Database (Oxford). 2017 Jan 1;2017(1):bax009. doi: 10.1093/database/bax009. Database (Oxford). 2017. PMID: 28415075 Free PMC article.
References
-
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

