The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species
- PMID: 26864432
- PMCID: PMC4771370
- DOI: 10.1128/MMBR.00040-15
The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species
Abstract
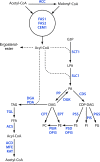
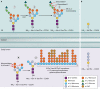
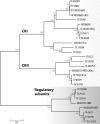
The genus Trichoderma contains fungi with high relevance for humans, with applications in enzyme production for plant cell wall degradation and use in biocontrol. Here, we provide a broad, comprehensive overview of the genomic content of these species for "hot topic" research aspects, including CAZymes, transport, transcription factors, and development, along with a detailed analysis and annotation of less-studied topics, such as signal transduction, genome integrity, chromatin, photobiology, or lipid, sulfur, and nitrogen metabolism in T. reesei, T. atroviride, and T. virens, and we open up new perspectives to those topics discussed previously. In total, we covered more than 2,000 of the predicted 9,000 to 11,000 genes of each Trichoderma species discussed, which is >20% of the respective gene content. Additionally, we considered available transcriptome data for the annotated genes. Highlights of our analyses include overall carbohydrate cleavage preferences due to the different genomic contents and regulation of the respective genes. We found light regulation of many sulfur metabolic genes. Additionally, a new Golgi 1,2-mannosidase likely involved in N-linked glycosylation was detected, as were indications for the ability of Trichoderma spp. to generate hybrid galactose-containing N-linked glycans. The genomic inventory of effector proteins revealed numerous compounds unique to Trichoderma, and these warrant further investigation. We found interesting expansions in the Trichoderma genus in several signaling pathways, such as G-protein-coupled receptors, RAS GTPases, and casein kinases. A particularly interesting feature absolutely unique to T. atroviride is the duplication of the alternative sulfur amino acid synthesis pathway.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
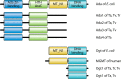
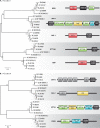
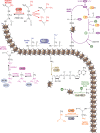
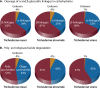


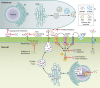
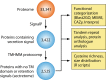
References
-
- Gupta VK, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy MG. 2014. Biotechnology and biology of Trichoderma. Elsevier, Oxford, United Kingdom.
-
- Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M. 2013. Trichoderma: biology and applications. CAB International, Oxfordshire, United Kingdom.
-
- Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, McCluskey K, Coulpier F, Deshpande N, von Dohren H, Ebbole DJ, Esquivel-Naranjo EU, Fekete E, Flipphi M, Glaser F, Gomez-Rodriguez EY, Gruber S, Han C, Henrissat B, Hermosa R, Hernandez-Onate M, Karaffa L, Kosti I, Le Crom S, Lindquist E, Lucas S, Lubeck M, Lubeck PS, Margeot A, Metz B, Misra M, Nevalainen H, Omann M, Packer N, Perrone G, Uresti-Rivera EE, Salamov A, Schmoll M, Seiboth B, Shapiro H, Sukno S, Tamayo-Ramos JA, Tisch D, Wiest A, Wilkinson HH, Zhang M, Coutinho PM, Kenerley CM, Monte E, Baker SE, Grigoriev IV. 2011. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol 12:R40. doi: 10.1186/gb-2011-12-4-r40. - DOI - PMC - PubMed
-
- Schmoll M, Seiboth B, Druzhinina I, Kubicek CP. 2014. Genomics analysis of biocontrol species and industrial enzyme producers from the genus Trichoderma, p 233–266. In Esser K, Nowrousian M (ed), The Mycota XIII. Springer, Berlin, Germany.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

