Metabolic disruption identified in the Huntington's disease transgenic sheep model
- PMID: 26864449
- PMCID: PMC4749952
- DOI: 10.1038/srep20681
Metabolic disruption identified in the Huntington's disease transgenic sheep model
Abstract
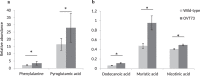
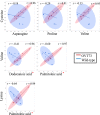
Huntington's disease (HD) is a dominantly inherited, progressive neurodegenerative disorder caused by a CAG repeat expansion within exon 1 of HTT, encoding huntingtin. There are no therapies that can delay the progression of this devastating disease. One feature of HD that may play a critical role in its pathogenesis is metabolic disruption. Consequently, we undertook a comparative study of metabolites in our transgenic sheep model of HD (OVT73). This model does not display overt symptoms of HD but has circadian rhythm alterations and molecular changes characteristic of the early phase disease. Quantitative metabolite profiles were generated from the motor cortex, hippocampus, cerebellum and liver tissue of 5 year old transgenic sheep and matched controls by gas chromatography-mass spectrometry. Differentially abundant metabolites were evident in the cerebellum and liver. There was striking tissue-specificity, with predominantly amino acids affected in the transgenic cerebellum and fatty acids in the transgenic liver, which together may indicate a hyper-metabolic state. Furthermore, there were more strong pair-wise correlations of metabolite abundance in transgenic than in wild-type cerebellum and liver, suggesting altered metabolic constraints. Together these differences indicate a metabolic disruption in the sheep model of HD and could provide insight into the presymptomatic human disease.
Figures

Similar articles
-
HdhQ111 Mice Exhibit Tissue Specific Metabolite Profiles that Include Striatal Lipid Accumulation.PLoS One. 2015 Aug 21;10(8):e0134465. doi: 10.1371/journal.pone.0134465. eCollection 2015. PLoS One. 2015. PMID: 26295712 Free PMC article.
-
Metabolomic Analysis of Plasma in Huntington's Disease Transgenic Sheep (Ovis aries) Reveals Progressive Circadian Rhythm Dysregulation.J Huntingtons Dis. 2023;12(1):31-42. doi: 10.3233/JHD-220552. J Huntingtons Dis. 2023. PMID: 36617787
-
Brain urea increase is an early Huntington's disease pathogenic event observed in a prodromal transgenic sheep model and HD cases.Proc Natl Acad Sci U S A. 2017 Dec 26;114(52):E11293-E11302. doi: 10.1073/pnas.1711243115. Epub 2017 Dec 11. Proc Natl Acad Sci U S A. 2017. PMID: 29229845 Free PMC article.
-
Hypothalamic alterations in Huntington's disease patients: comparison with genetic rodent models.J Neuroendocrinol. 2014 Nov;26(11):761-75. doi: 10.1111/jne.12190. J Neuroendocrinol. 2014. PMID: 25074766 Review.
-
Do Disruptions in the Circadian Timing System Contribute to Autonomic Dysfunction in Huntington's Disease?Yale J Biol Med. 2019 Jun 27;92(2):291-303. eCollection 2019 Jun. Yale J Biol Med. 2019. PMID: 31249490 Free PMC article. Review.
Cited by
-
Cerebellum in Alzheimer's disease and other neurodegenerative diseases: an emerging research frontier.MedComm (2020). 2024 Jul 13;5(7):e638. doi: 10.1002/mco2.638. eCollection 2024 Jul. MedComm (2020). 2024. PMID: 39006764 Free PMC article. Review.
-
From Pathogenesis to Therapeutics: A Review of 150 Years of Huntington's Disease Research.Int J Mol Sci. 2023 Aug 21;24(16):13021. doi: 10.3390/ijms241613021. Int J Mol Sci. 2023. PMID: 37629202 Free PMC article. Review.
-
Sheep recognize familiar and unfamiliar human faces from two-dimensional images.R Soc Open Sci. 2017 Nov 8;4(11):171228. doi: 10.1098/rsos.171228. eCollection 2017 Nov. R Soc Open Sci. 2017. PMID: 29291109 Free PMC article.
-
Metabolomics: An Emerging "Omics" Platform for Systems Biology and Its Implications for Huntington Disease Research.Metabolites. 2023 Dec 18;13(12):1203. doi: 10.3390/metabo13121203. Metabolites. 2023. PMID: 38132886 Free PMC article. Review.
-
Metabolism in Huntington's disease: a major contributor to pathology.Metab Brain Dis. 2022 Aug;37(6):1757-1771. doi: 10.1007/s11011-021-00844-y. Epub 2021 Oct 27. Metab Brain Dis. 2022. PMID: 34704220 Review.
References
-
- HDCRG. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971–983 (1993). - PubMed
-
- Hodges A. et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet 15, 965–77 (2006). - PubMed
-
- Underwood B. R. et al. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain 129, 877–886 (2006). - PubMed
-
- Mochel F., Benaich S., Rabier D. & Durr A. Validation of plasma branched chain amino acids as biomarkers in Huntington disease. Arch Neurol 68, 265–7 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

