New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases
- PMID: 26865263
- PMCID: PMC4749974
- DOI: 10.1038/srep20629
New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases
Erratum in
-
Erratum: New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases.Sci Rep. 2016 Apr 20;6:23879. doi: 10.1038/srep23879. Sci Rep. 2016. PMID: 27094544 Free PMC article. No abstract available.
Abstract
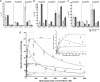
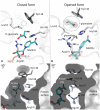
Glyoxylate accumulation within cells is highly toxic. In humans, it is associated with hyperoxaluria type 2 (PH2) leading to renal failure. The glyoxylate content within cells is regulated by the NADPH/NADH dependent glyoxylate/hydroxypyruvate reductases (GRHPR). These are highly conserved enzymes with a dual activity as they are able to reduce glyoxylate to glycolate and to convert hydroxypyruvate into D-glycerate. Despite the determination of high-resolution X-ray structures, the substrate recognition mode of this class of enzymes remains unclear. We determined the structure at 2.0 Å resolution of a thermostable GRHPR from Archaea as a ternary complex in the presence of D-glycerate and NADPH. This shows a binding mode conserved between human and archeal enzymes. We also determined the first structure of GRHPR in presence of glyoxylate at 1.40 Å resolution. This revealed the pivotal role of Leu53 and Trp138 in substrate trafficking. These residues act as gatekeepers at the entrance of a tunnel connecting the active site to protein surface. Taken together, these results allowed us to propose a general model for GRHPR mode of action.
Figures


References
-
- Yoshikawa S. et al. Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate. Acta Crystallogr. D Biol. Crystallogr. 63, 357–365 (2007). - PubMed
-
- Rumsby G. & Cregeen D. P. Identification and expression of a cDNA for human hydroxypyruvate/glyoxylate reductase. Biochimica et Biophysica Acta 1446, 383–388 (1999). - PubMed
-
- Hullin R. P. Glyoxylate reductase, two forms from Pseudomonas. Meth. Enzymol. 41, 343–348 (1975). - PubMed
-
- Yoshida T. et al. Cloning and expression of the gene for hydroxypyruvate reductase (D-glycerate dehydrogenase from an obligate methylotroph Hyphomicrobium methylovorum GM2. Eur. J. Biochem. 223, 727–732 (1994). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

