A Highly Conserved Salt Bridge Stabilizes the Kinked Conformation of β2,3-Sheet Essential for Channel Function of P2X4 Receptors
- PMID: 26865631
- PMCID: PMC4825005
- DOI: 10.1074/jbc.M115.711127
A Highly Conserved Salt Bridge Stabilizes the Kinked Conformation of β2,3-Sheet Essential for Channel Function of P2X4 Receptors
Abstract
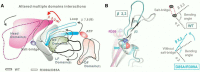
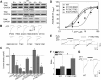
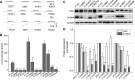
Significant progress has been made in understanding the roles of crucial residues/motifs in the channel function of P2X receptors during the pre-structure era. The recent structural determination of P2X receptors allows us to reevaluate the role of those residues/motifs. Residues Arg-309 and Asp-85 (rat P2X4 numbering) are highly conserved throughout the P2X family and were involved in loss-of-function polymorphism in human P2X receptors. Previous studies proposed that they participated in direct ATP binding. However, the crystal structure of P2X demonstrated that those two residues form an intersubunit salt bridge located far away from the ATP-binding site. Therefore, it is necessary to reevaluate the role of this salt bridge in P2X receptors. Here, we suggest the crucial role of this structural element both in protein stability and in channel gating rather than direct ATP interaction and channel assembly. Combining mutagenesis, charge swap, and disulfide cross-linking, we revealed the stringent requirement of this salt bridge in normal P2X4 channel function. This salt bridge may contribute to stabilizing the bending conformation of the β2,3-sheet that is structurally coupled with this salt bridge and the α2-helix. Strongly kinked β2,3 is essential for domain-domain interactions between head domain, dorsal fin domain, right flipper domain, and loop β7,8 in P2X4 receptors. Disulfide cross-linking with directions opposing or along the bending angle of the β2,3-sheet toward the α2-helix led to loss-of-function and gain-of-function of P2X4 receptors, respectively. Further insertion of amino acids with bulky side chains into the linker between the β2,3-sheet or the conformational change of the α2-helix, interfering with the kinked conformation of β2,3, led to loss-of-function of P2X4 receptors. All these findings provided new insights in understanding the contribution of the salt bridge between Asp-85 and Arg-309 and its structurally coupled β2,3-sheet to the function of P2X receptors.
Keywords: ATP; P2X receptors; conformational change; ion channel; protein expression; protein stability; salt bridge.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Intersubunit physical couplings fostered by the left flipper domain facilitate channel opening of P2X4 receptors.J Biol Chem. 2017 May 5;292(18):7619-7635. doi: 10.1074/jbc.M116.771121. Epub 2017 Mar 16. J Biol Chem. 2017. PMID: 28302727 Free PMC article.
-
Identification of functionally important residues of the rat P2X4 receptor by alanine scanning mutagenesis of the dorsal fin and left flipper domains.PLoS One. 2014 Nov 14;9(11):e112902. doi: 10.1371/journal.pone.0112902. eCollection 2014. PLoS One. 2014. PMID: 25398027 Free PMC article.
-
Inherent dynamics of head domain correlates with ATP-recognition of P2X4 receptors: insights gained from molecular simulations.PLoS One. 2014 May 30;9(5):e97528. doi: 10.1371/journal.pone.0097528. eCollection 2014. PLoS One. 2014. PMID: 24878662 Free PMC article.
-
Structural interpretation of P2X receptor mutagenesis studies on drug action.Br J Pharmacol. 2010 Nov;161(5):961-71. doi: 10.1111/j.1476-5381.2010.00728.x. Br J Pharmacol. 2010. PMID: 20977449 Free PMC article. Review.
-
Structural insights into the function of P2X4: an ATP-gated cation channel of neuroendocrine cells.Cell Mol Neurobiol. 2010 Nov;30(8):1251-8. doi: 10.1007/s10571-010-9568-y. Epub 2010 Nov 25. Cell Mol Neurobiol. 2010. PMID: 21107680 Free PMC article. Review.
Cited by
-
PD-L1 Nanobody Competitively Inhibits the Formation of the PD-1/PD-L1 Complex: Comparative Molecular Dynamics Simulations.Int J Mol Sci. 2018 Jul 7;19(7):1984. doi: 10.3390/ijms19071984. Int J Mol Sci. 2018. PMID: 29986511 Free PMC article.
-
Identification of the molecular determinants of antagonist potency in the allosteric binding pocket of human P2X4.Front Pharmacol. 2023 Feb 9;14:1101023. doi: 10.3389/fphar.2023.1101023. eCollection 2023. Front Pharmacol. 2023. PMID: 36843952 Free PMC article.
-
The long β2,3-sheets encoded by redundant sequences play an integral role in the channel function of P2X7 receptors.J Biol Chem. 2022 Jun;298(6):102002. doi: 10.1016/j.jbc.2022.102002. Epub 2022 Apr 30. J Biol Chem. 2022. PMID: 35504351 Free PMC article.
-
Mapping a novel positive allosteric modulator binding site in the central vestibule region of human P2X7.Sci Rep. 2019 Mar 1;9(1):3231. doi: 10.1038/s41598-019-39771-5. Sci Rep. 2019. PMID: 30824738 Free PMC article.
-
Intersubunit physical couplings fostered by the left flipper domain facilitate channel opening of P2X4 receptors.J Biol Chem. 2017 May 5;292(18):7619-7635. doi: 10.1074/jbc.M116.771121. Epub 2017 Mar 16. J Biol Chem. 2017. PMID: 28302727 Free PMC article.
References
-
- Surprenant A., and North R. A. (2009) Signaling at purinergic P2X receptors. Annu. Rev. Physiol. 71, 333–359 - PubMed
-
- Khakh B. S., and North R. A. (2006) P2X receptors as cell-surface ATP sensors in health and disease. Nature 442, 527–532 - PubMed
-
- Lewis C., Neidhart S., Holy C., North R. A., Buell G., and Surprenant A. (1995) Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature 377, 432–435 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

