Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses
- PMID: 26865708
- PMCID: PMC4810531
- DOI: 10.1128/JVI.02804-15
Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses
Abstract
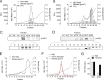
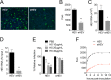
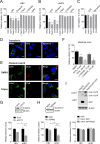
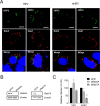
The hepatitis E virus (HEV) sheds into feces as nonenveloped virions but circulates in the blood in a membrane-associated, quasi-enveloped form (eHEV). Since the eHEV virions lack viral proteins on the surface, we investigated the entry mechanism for eHEV. We found that compared to nonenveloped HEV virions, eHEV attachment to the cell was much less efficient, requiring a longer inoculation time to reach its maximal infectivity. A survey of cellular internalization pathways identified clathrin-mediated endocytosis as the main route for eHEV entry. Unlike nonenveloped HEV virions, eHEV entry requires Rab5 and Rab7, small GTPases involved in endosomal trafficking, and blocking endosomal acidification abrogated eHEV infectivity. However, low pH alone was not sufficient for eHEV uncoating, suggesting that additional steps are required for entry. Supporting this concept, eHEV infectivity was substantially reduced in cells depleted of Niemann-Pick disease type C1, a lysosomal protein required for cholesterol extraction from lipid, or in cells treated with an inhibitor of lysosomal acid lipase. These data support a model in which the quasi-envelope is degraded within the lysosome prior to virus uncoating, a potentially novel mechanism for virus entry.
Importance: The recent discovery of quasi-enveloped viruses has shifted the paradigm of virus-host interactions. The impact of quasi-envelopment in the virus life cycle and pathogenesis is largely unknown. HEV is a highly relevant model to study these questions. HEV circulates as quasi-enveloped virions in the blood that are hidden from neutralizing antibodies. eHEV particles most likely are responsible for the cell-to-cell spread of the virus. Given the increasing concerns about persistent HEV infection and its potential for transmission via the blood supply, understanding how eHEV infects cells is important for understanding its pathogenesis and developing therapies. Our data provide evidence that eHEV uses a potentially novel mechanism for cellular entry. Several steps critical to eHEV entry were identified and may provide a basis for developing treatments for hepatitis E. Because quasi-enveloped viruses resemble exosomes, these data also may provide insights into the exosome-mediated intercellular communications.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Hepatitis E Virus Entry.Viruses. 2019 Sep 20;11(10):883. doi: 10.3390/v11100883. Viruses. 2019. PMID: 31547135 Free PMC article. Review.
-
Integrin beta 1 facilitates non-enveloped hepatitis E virus cell entry through the recycling endosome.Nat Commun. 2025 Jun 26;16(1):5403. doi: 10.1038/s41467-025-61071-y. Nat Commun. 2025. PMID: 40571699 Free PMC article.
-
The different replication between nonenveloped and quasi-enveloped hepatitis E virus.J Med Virol. 2021 Nov;93(11):6267-6277. doi: 10.1002/jmv.27121. Epub 2021 Jun 10. J Med Virol. 2021. PMID: 34076903
-
Role of Rab13, Protein Kinase A, and Zonula Occludens-1 in Hepatitis E Virus Entry and Cell-to-Cell Spread: Comparative Analysis of Quasi-Enveloped and Non-Enveloped Forms.Pathogens. 2024 Dec 20;13(12):1130. doi: 10.3390/pathogens13121130. Pathogens. 2024. PMID: 39770389 Free PMC article.
-
Hepatitis E Virus: What More Do We Need to Know?Medicina (Kaunas). 2024 Jun 18;60(6):998. doi: 10.3390/medicina60060998. Medicina (Kaunas). 2024. PMID: 38929615 Free PMC article. Review.
Cited by
-
Expression Profiles of Exosomal MicroRNAs from HEV- and HCV-Infected Blood Donors and Patients: A Pilot Study.Viruses. 2020 Jul 30;12(8):833. doi: 10.3390/v12080833. Viruses. 2020. PMID: 32751663 Free PMC article.
-
Hepatitis E Virus Entry.Viruses. 2019 Sep 20;11(10):883. doi: 10.3390/v11100883. Viruses. 2019. PMID: 31547135 Free PMC article. Review.
-
Cell Culture Models for Hepatitis E Virus.Viruses. 2019 Jul 3;11(7):608. doi: 10.3390/v11070608. Viruses. 2019. PMID: 31277308 Free PMC article. Review.
-
Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection.Proc Natl Acad Sci U S A. 2018 May 1;115(18):4773-4778. doi: 10.1073/pnas.1721345115. Epub 2018 Apr 18. Proc Natl Acad Sci U S A. 2018. PMID: 29669922 Free PMC article.
-
Cathepsins in cellular entry of human pathogenic viruses.J Virol. 2025 Apr 15;99(4):e0164224. doi: 10.1128/jvi.01642-24. Epub 2025 Mar 26. J Virol. 2025. PMID: 40135892 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

