Hepatitis B Virus Infection of a Mouse Hepatic Cell Line Reconstituted with Human Sodium Taurocholate Cotransporting Polypeptide
- PMID: 26865711
- PMCID: PMC4836309
- DOI: 10.1128/JVI.02832-15
Hepatitis B Virus Infection of a Mouse Hepatic Cell Line Reconstituted with Human Sodium Taurocholate Cotransporting Polypeptide
Abstract
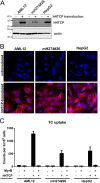
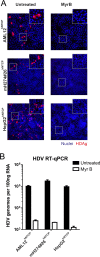
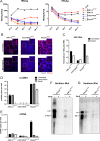
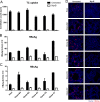
Hepatitis B virus (HBV) enters hepatocytes via its receptor, human sodium taurocholate cotransporting polypeptide (hNTCP). So far, HBV infection has been achieved only in human hepatic cells reconstituted with hNTCP and not in cells of mouse origin. Here, the first mouse liver cell line (AML12) which gains susceptibility to HBV upon hNTCP expression is described. Thus, HBV infection of receptor-expressing mouse hepatocytes does not principally require a human cofactor but can be triggered by endogenous murine determinants.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques.Nat Commun. 2017 Dec 15;8(1):2146. doi: 10.1038/s41467-017-01953-y. Nat Commun. 2017. PMID: 29247188 Free PMC article.
-
Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide.J Virol. 2014 Mar;88(6):3273-84. doi: 10.1128/JVI.03478-13. Epub 2014 Jan 3. J Virol. 2014. PMID: 24390325 Free PMC article.
-
Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes.Hepatology. 2017 Sep;66(3):703-716. doi: 10.1002/hep.29112. Epub 2017 Jul 18. Hepatology. 2017. PMID: 28195359
-
Molecular regulation of the hepatic bile acid uptake transporter and HBV entry receptor NTCP.Biochim Biophys Acta Mol Cell Biol Lipids. 2021 Aug;1866(8):158960. doi: 10.1016/j.bbalip.2021.158960. Epub 2021 Apr 29. Biochim Biophys Acta Mol Cell Biol Lipids. 2021. PMID: 33932583 Review.
-
NTCP opens the door for hepatitis B virus infection.Antiviral Res. 2015 Sep;121:24-30. doi: 10.1016/j.antiviral.2015.06.002. Epub 2015 Jun 10. Antiviral Res. 2015. PMID: 26071008 Review.
Cited by
-
Preclinical assessment of antiviral combination therapy in a genetically humanized mouse model for hepatitis delta virus infection.Sci Transl Med. 2018 Jun 27;10(447):eaap9328. doi: 10.1126/scitranslmed.aap9328. Sci Transl Med. 2018. PMID: 29950446 Free PMC article.
-
Human apolipoprotein E promotes hepatitis B virus infection and production.PLoS Pathog. 2019 Aug 8;15(8):e1007874. doi: 10.1371/journal.ppat.1007874. eCollection 2019 Aug. PLoS Pathog. 2019. PMID: 31393946 Free PMC article.
-
Cell and Animal Models for Studying Hepatitis B Virus Infection and Drug Development.Gastroenterology. 2019 Jan;156(2):338-354. doi: 10.1053/j.gastro.2018.06.093. Epub 2018 Sep 19. Gastroenterology. 2019. PMID: 30243619 Free PMC article. Review.
-
Region-Specific Hepatitis B Virus Genome Exposure from Nucleocapsid Modulated by Capsid Linker Sequence and Inhibitor: Implications for Uncoating.J Virol. 2022 Apr 27;96(8):e0039922. doi: 10.1128/jvi.00399-22. Epub 2022 Apr 7. J Virol. 2022. PMID: 35389266 Free PMC article.
-
DNA Polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA.PLoS Pathog. 2019 Apr 26;15(4):e1007742. doi: 10.1371/journal.ppat.1007742. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 31026293 Free PMC article.
References
-
- Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi:10.1053/j.gastro.2013.12.024. - DOI - PubMed
-
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. doi:10.7554/eLife.00049. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

