Role of TSPAN9 in Alphavirus Entry and Early Endosomes
- PMID: 26865714
- PMCID: PMC4836341
- DOI: 10.1128/JVI.00018-16
Role of TSPAN9 in Alphavirus Entry and Early Endosomes
Abstract
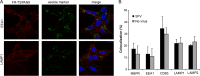
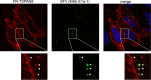
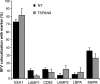
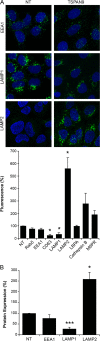
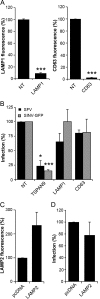
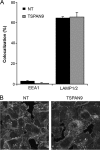
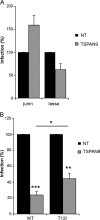
Alphaviruses are small enveloped RNA viruses that infect cells via clathrin-mediated endocytosis and low-pH-triggered fusion in the early endosome. Using a small interfering RNA (siRNA) screen in human cells, we previously identified TSPAN9 as a host factor that promotes infection by the alphaviruses Sindbis virus (SINV), Semliki Forest virus (SFV), and chikungunya virus (CHIKV). Depletion of TSPAN9 specifically decreases SFV membrane fusion in endosomes. TSPAN9 is a member of the tetraspanin family of multipass membrane proteins, but its cellular function is currently unknown. Here we used U-2 OS cells stably overexpressing TSPAN9 to show that TSPAN9 is localized at the plasma membrane and in early and late endosomes. Internalized SFV particles colocalized with TSPAN9 in vesicles early during infection. Depletion of TSPAN9 led to reductions in the amounts of the late endosomal proteins LAMP1 and CD63 and an increase in the amount of LAMP2. However, TSPAN9 depletion did not alter the delivery of SFV to early endosomes or change their pH or protease activity. Comparative studies showed that TSPAN9 depletion strongly inhibited infection by several viruses that fuse in early endosomes (SFV, SINV, CHIKV, and vesicular stomatitis virus [VSV]), while viruses that fuse in the late endosome (recombinant VSV-Lassa and VSV-Junin), including an SFV point mutant with a lower pH threshold for fusion (SFV E2 T12I), were relatively resistant. Our data suggest that TSPAN9 modulates the early endosome compartment to make it more permissive for membrane fusion of early-penetrating viruses.
Importance: Alphaviruses are spread by mosquitoes and can cause serious human diseases such as arthritis and encephalitis. Recent outbreaks of CHIKV infection are responsible for millions of cases of acute illness and long-term complications. There are no vaccines or antiviral treatments for these important human pathogens. Alphaviruses infect host cells by utilizing the endocytic machinery of the cell and fusing their membrane with that of the endosome. Although the mechanism of virus-membrane fusion is well studied, we still know relatively little about the host cell proteins that are involved in alphavirus entry. Here we characterized the role of the host membrane protein TSPAN9 in alphavirus infection. TSPAN9 was localized to early endosomes containing internalized alphavirus, and depletion of TSPAN9 inhibited virus fusion with the early endosome membrane. In contrast, infection of viruses that enter through the late endosome was relatively resistant to TSPAN9 depletion, suggesting an important role for TSPAN9 in the early endosome.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
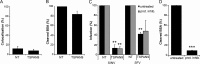
Similar articles
-
Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry.PLoS Pathog. 2013;9(12):e1003835. doi: 10.1371/journal.ppat.1003835. Epub 2013 Dec 19. PLoS Pathog. 2013. PMID: 24367265 Free PMC article.
-
Alphavirus Restriction by IFITM Proteins.Traffic. 2016 Sep;17(9):997-1013. doi: 10.1111/tra.12416. Epub 2016 Jun 24. Traffic. 2016. PMID: 27219333 Free PMC article.
-
Dissecting the Role of E2 Protein Domains in Alphavirus Pathogenicity.J Virol. 2015 Dec 16;90(5):2418-33. doi: 10.1128/JVI.02792-15. J Virol. 2015. PMID: 26676771 Free PMC article.
-
Alphavirus entry into host cells.Prog Mol Biol Transl Sci. 2015;129:33-62. doi: 10.1016/bs.pmbts.2014.10.002. Epub 2014 Dec 1. Prog Mol Biol Transl Sci. 2015. PMID: 25595800 Review.
-
The Alphavirus Exit Pathway: What We Know and What We Wish We Knew.Viruses. 2018 Feb 22;10(2):89. doi: 10.3390/v10020089. Viruses. 2018. PMID: 29470397 Free PMC article. Review.
Cited by
-
The Tetraspanin CD81 Is a Host Factor for Chikungunya Virus Replication.mBio. 2022 Jun 28;13(3):e0073122. doi: 10.1128/mbio.00731-22. Epub 2022 May 25. mBio. 2022. PMID: 35612284 Free PMC article.
-
Role of the vacuolar ATPase in the Alphavirus replication cycle.Heliyon. 2018 Jul 25;4(7):e00701. doi: 10.1016/j.heliyon.2018.e00701. eCollection 2018 Jul. Heliyon. 2018. PMID: 30094371 Free PMC article.
-
Genome-Wide Approaches to Unravel the Host Factors Involved in Chikungunya Virus Replication.Front Microbiol. 2022 Mar 24;13:866271. doi: 10.3389/fmicb.2022.866271. eCollection 2022. Front Microbiol. 2022. PMID: 35401487 Free PMC article. Review.
-
TSPAN9 suppresses the chemosensitivity of gastric cancer to 5-fluorouracil by promoting autophagy.Cancer Cell Int. 2020 Jan 3;20:4. doi: 10.1186/s12935-019-1089-2. eCollection 2020. Cancer Cell Int. 2020. PMID: 31911756 Free PMC article.
-
Oncolytic alphavirus-induced extracellular vesicles counteract the immunosuppressive effect of melanoma-derived extracellular vesicles.Sci Rep. 2025 Jan 4;15(1):803. doi: 10.1038/s41598-024-82331-9. Sci Rep. 2025. PMID: 39755711 Free PMC article.
References
-
- Kuhn RJ. 2013. Togaviridae, p 629–650. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

