The Autographa californica Multiple Nucleopolyhedrovirus ac54 Gene Is Crucial for Localization of the Major Capsid Protein VP39 at the Site of Nucleocapsid Assembly
- PMID: 26865720
- PMCID: PMC4810565
- DOI: 10.1128/JVI.02885-15
The Autographa californica Multiple Nucleopolyhedrovirus ac54 Gene Is Crucial for Localization of the Major Capsid Protein VP39 at the Site of Nucleocapsid Assembly
Abstract
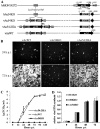
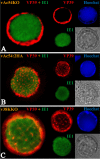
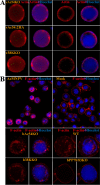
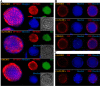
Baculovirus DNAs are synthesized and inserted into preformed capsids to form nucleocapsids at a site in the infected cell nucleus, termed the virogenic stroma. Nucleocapsid assembly ofAutographa californicamultiple nucleopolyhedrovirus (AcMNPV) requires the major capsid protein VP39 and nine minor capsid proteins, including VP1054. However, how VP1054 participates in nucleocapsid assembly remains elusive. In this study, the VP1054-encoding gene (ac54) was deleted to generate theac54-knockout AcMNPV (vAc54KO). In vAc54KO-transfected cells, nucleocapsid assembly was disrupted, leading to the formation of abnormally elongated capsid structures. Interestingly, unlike cells transfected with AcMNPV mutants lacking other minor capsid proteins, in which capsid structures were distributed within the virogenic stroma,ac54ablation resulted in a distinctive location of capsid structures and VP39 at the periphery of the nucleus. The altered distribution pattern of capsid structures was also observed in cells transfected with AcMNPV lacking BV/ODV-C42 or in cytochalasind-treated AcMNPV-infected cells. BV/ODV-C42, along with PP78/83, has been shown to promote nuclear filamentous actin (F-actin) formation, which is another requisite for nucleocapsid assembly. Immunofluorescence using phalloidin indicated that the formation and distribution of nuclear F-actin were not affected byac54deletion. However, immunoelectron microscopy revealed that BV/ODV-C42, PP78/83, and 38K failed to integrate into capsid structures in the absence of VP1054, and immunoprecipitation further demonstrated that in transient expression assays, VP1054 interacted with BV/ODV-C42 and VP80 but not VP39. Our findings suggest that VP1054 plays an important role in the transport of capsid proteins to the nucleocapsid assembly site prior to the process of nucleocapsid assembly.
Importance: Baculoviruses are large DNA viruses whose replication occurs within the host nucleus. The localization of capsids into the capsid assembly site requires virus-induced nuclear F-actin; the inhibition of nuclear F-actin formation results in the retention of capsid structures at the periphery of the nucleus. In this paper, we note that the minor capsid protein VP1054 is essential for the localization of capsid structures, the major capsid protein VP39, and the minor capsid protein 38K into the capsid assembly site. Moreover, VP1054 is crucial for correct targeting of the nuclear F-actin factors BV/ODV-C42 and PP78/83 for capsid maturation. However, the formation and distribution of nuclear F-actin are not affected by the lack of VP1054. We further reveal that VP1054 interacts with BV/ODV-C42 and a capsid transport-related protein, VP80. Taken together, our findings suggest that VP1054 plays a unique role in the pathway(s) for transport of capsid proteins.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Autographa californica multiple nucleopolyhedrovirus 38K is a novel nucleocapsid protein that interacts with VP1054, VP39, VP80, and itself.J Virol. 2008 Dec;82(24):12356-64. doi: 10.1128/JVI.00948-08. Epub 2008 Oct 15. J Virol. 2008. PMID: 18922869 Free PMC article.
-
Ac102 Participates in Nuclear Actin Polymerization by Modulating BV/ODV-C42 Ubiquitination during Autographa californica Multiple Nucleopolyhedrovirus Infection.J Virol. 2018 May 29;92(12):e00005-18. doi: 10.1128/JVI.00005-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618641 Free PMC article.
-
Autographa californica Nucleopolyhedrovirus AC141 (Exon0), a Potential E3 Ubiquitin Ligase, Interacts with Viral Ubiquitin and AC66 To Facilitate Nucleocapsid Egress.J Virol. 2018 Jan 17;92(3):e01713-17. doi: 10.1128/JVI.01713-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142135 Free PMC article.
-
me53 encoded by Autographa californica multiple nucleopolyhedrovirus: from mechanism to function.Virus Genes. 2023 Apr;59(2):188-194. doi: 10.1007/s11262-022-01943-3. Epub 2022 Oct 13. Virus Genes. 2023. PMID: 36229721 Review.
-
Nucleocapsid Assembly of Baculoviruses.Viruses. 2019 Jul 1;11(7):595. doi: 10.3390/v11070595. Viruses. 2019. PMID: 31266177 Free PMC article. Review.
Cited by
-
The 38K-Mediated Specific Dephosphorylation of the Viral Core Protein P6.9 Plays an Important Role in the Nucleocapsid Assembly of Autographa californica Multiple Nucleopolyhedrovirus.J Virol. 2018 Apr 13;92(9):e01989-17. doi: 10.1128/JVI.01989-17. Print 2018 May 1. J Virol. 2018. PMID: 29444944 Free PMC article.
-
The Autographa californica Multiple Nucleopolyhedrovirus ac83 Gene Contains a cis-Acting Element That Is Essential for Nucleocapsid Assembly.J Virol. 2017 Feb 14;91(5):e02110-16. doi: 10.1128/JVI.02110-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28031366 Free PMC article.
-
VP39 of Spodoptera litura multicapsid nucleopolyhedrovirus cannot efficiently rescue the nucleocapsid assembly of vp39-null Autographa californica multiple nucleopolyhedrovirus.Virol J. 2021 Apr 20;18(1):81. doi: 10.1186/s12985-021-01553-9. Virol J. 2021. PMID: 33879205 Free PMC article.
-
Architecture of the baculovirus nucleocapsid revealed by cryo-EM.Nat Commun. 2023 Nov 18;14(1):7481. doi: 10.1038/s41467-023-43284-1. Nat Commun. 2023. PMID: 37980340 Free PMC article.
-
Nuclear Cytoskeleton in Virus Infection.Int J Mol Sci. 2022 Jan 5;23(1):578. doi: 10.3390/ijms23010578. Int J Mol Sci. 2022. PMID: 35009004 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

