Genome-Wide Association Study in Arabidopsis thaliana of Natural Variation in Seed Oil Melting Point: A Widespread Adaptive Trait in Plants
- PMID: 26865732
- PMCID: PMC4885241
- DOI: 10.1093/jhered/esw008
Genome-Wide Association Study in Arabidopsis thaliana of Natural Variation in Seed Oil Melting Point: A Widespread Adaptive Trait in Plants
Abstract
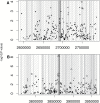
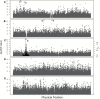
Seed oil melting point is an adaptive, quantitative trait determined by the relative proportions of the fatty acids that compose the oil. Micro- and macro-evolutionary evidence suggests selection has changed the melting point of seed oils to covary with germination temperatures because of a trade-off between total energy stores and the rate of energy acquisition during germination under competition. The seed oil compositions of 391 natural accessions of Arabidopsis thaliana, grown under common-garden conditions, were used to assess whether seed oil melting point within a species varied with germination temperature. In support of the adaptive explanation, long-term monthly spring and fall field temperatures of the accession collection sites significantly predicted their seed oil melting points. In addition, a genome-wide association study (GWAS) was performed to determine which genes were most likely responsible for the natural variation in seed oil melting point. The GWAS found a single highly significant association within the coding region of FAD2, which encodes a fatty acid desaturase central to the oil biosynthesis pathway. In a separate analysis of 15 a priori oil synthesis candidate genes, 2 (FAD2 and FATB) were located near significant SNPs associated with seed oil melting point. These results comport with others' molecular work showing that lines with alterations in these genes affect seed oil melting point as expected. Our results suggest natural selection has acted on a small number of loci to alter a quantitative trait in response to local environmental conditions.
Keywords: GWAS.; adaptation; evolution; fatty acid composition.
© The American Genetic Association. 2016. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Genome-Wide Association Study of Arabidopsis thaliana Identifies Determinants of Natural Variation in Seed Oil Composition.J Hered. 2016 May;107(3):248-56. doi: 10.1093/jhered/esv100. Epub 2015 Dec 24. J Hered. 2016. PMID: 26704140 Free PMC article.
-
Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana.Plant J. 2013 Jun;74(5):815-28. doi: 10.1111/tpj.12167. Epub 2013 Apr 4. Plant J. 2013. PMID: 23464703
-
Natural variation for seed oil composition in Arabidopsis thaliana.Phytochemistry. 2003 Nov;64(6):1077-90. doi: 10.1016/s0031-9422(03)00351-0. Phytochemistry. 2003. PMID: 14568074
-
Emergence timing and fitness consequences of variation in seed oil composition in Arabidopsis thaliana.Ecol Evol. 2015 Jan;5(1):164-71. doi: 10.1002/ece3.1265. Epub 2014 Dec 10. Ecol Evol. 2015. PMID: 25628873 Free PMC article.
-
When Size Matters: New Insights on How Seed Size Can Contribute to the Early Stages of Plant Development.Plants (Basel). 2024 Jun 28;13(13):1793. doi: 10.3390/plants13131793. Plants (Basel). 2024. PMID: 38999633 Free PMC article. Review.
Cited by
-
Genome Wide Analysis of Fatty Acid Desaturation and Its Response to Temperature.Plant Physiol. 2017 Mar;173(3):1594-1605. doi: 10.1104/pp.16.01907. Epub 2017 Jan 20. Plant Physiol. 2017. PMID: 28108698 Free PMC article.
-
The genetic architecture of amylose biosynthesis in maize kernel.Plant Biotechnol J. 2018 Feb;16(2):688-695. doi: 10.1111/pbi.12821. Epub 2017 Sep 15. Plant Biotechnol J. 2018. PMID: 28796926 Free PMC article.
-
Extensive simulations assess the performance of genome-wide association mapping in various Saccharomyces cerevisiae subpopulations.Philos Trans R Soc Lond B Biol Sci. 2022 Jul 18;377(1855):20200514. doi: 10.1098/rstb.2020.0514. Epub 2022 May 30. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35634920 Free PMC article.
-
Genome-wide association study reveals candidate genes influencing lipids and diterpenes contents in Coffea arabica L.Sci Rep. 2018 Jan 11;8(1):465. doi: 10.1038/s41598-017-18800-1. Sci Rep. 2018. PMID: 29323254 Free PMC article.
-
Correlations of Genotype with Climate Parameters Suggest Caenorhabditis elegans Niche Adaptations.G3 (Bethesda). 2017 Jan 5;7(1):289-298. doi: 10.1534/g3.116.035162. G3 (Bethesda). 2017. PMID: 27866149 Free PMC article.
References
-
- Anastasio AE, Platt A, Horton M, Grotewold E, Scholl R, Borevitz JO, Nordborg M, Bergelson J. 2011. Source verification of mis-identified Arabidopsis thaliana accessions. Plant J. 67:554–566. - PubMed
-
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. - PubMed
-
- Beck JB, Schmuths H, Schaal BA. 2008. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol. 17:902–915. - PubMed
-
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57:289–300.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

